













| General Information: |
|
| Name(s) found: |
KRE33 /
YNL132W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1056 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
ribosome biogenesis
[RCA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKKAIDSRI PSLIRNGVQT KQRSIFVIVG DRARNQLPNL HYLMMSADLK MNKSVLWAYK 60
61 KKLLGFTSHR KKRENKIKKE IKRGTREVNE MDPFESFISN QNIRYVYYKE SEKILGNTYG 120
121 MCILQDFEAL TPNLLARTIE TVEGGGIVVI LLKSMSSLKQ LYTMTMDVHA RYRTEAHGDV 180
181 VARFNERFIL SLGSNPNCLV VDDELNVLPL SGAKNVKPLP PKEDDELPPK QLELQELKES 240
241 LEDVQPAGSL VSLSKTVNQA HAILSFIDAI SEKTLNFTVA LTAGRGRGKS AALGISIAAA 300
301 VSHGYSNIFV TSPSPENLKT LFEFIFKGFD ALGYQEHIDY DIIQSTNPDF NKAIVRVDIK 360
361 RDHRQTIQYI VPQDHQVLGQ AELVVIDEAA AIPLPIVKNL LGPYLVFMAS TINGYEGTGR 420
421 SLSLKLIQQL RNQNNTSGRE STQTAVVSRD NKEKDSHLHS QSRQLREISL DEPIRYAPGD 480
481 PIEKWLNKLL CLDVTLIKNP RFATRGTPHP SQCNLFVVNR DTLFSYHPVS ENFLEKMMAL 540
541 YVSSHYKNSP NDLQLMSDAP AHKLFVLLPP IDPKDGGRIP DPLCVIQIAL EGEISKESVR 600
601 NSLSRGQRAG GDLIPWLISQ QFQDEEFASL SGARIVRIAT NPEYASMGYG SRAIELLRDY 660
661 FEGKFTDMSE DVRPKDYSIK RVSDKELAKT NLLKDDVKLR DAKTLPPLLL KLSEQPPHYL 720
721 HYLGVSYGLT QSLHKFWKNN SFVPVYLRQT ANDLTGEHTC VMLNVLEGRE SNWLVEFAKD 780
781 FRKRFLSLLS YDFHKFTAVQ ALSVIESSKK AQDLSDDEKH DNKELTRTHL DDIFSPFDLK 840
841 RLDSYSNNLL DYHVIGDMIP MLALLYFGDK MGDSVKLSSV QSAILLAIGL QRKNIDTIAK 900
901 ELNLPSNQTI AMFAKIMRKM SQYFRQLLSQ SIEETLPNIK DDAIAEMDGE EIKNYNAAEA 960
961 LDQMEEDLEE AGSEAVQAMR EKQKELINSL NLDKYAINDN SEEWAESQKS LEIAAKAKGV1020
1021 VSLKTGKKRT TEKAEDIYRQ EMKAMKKPRK SKKAAN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
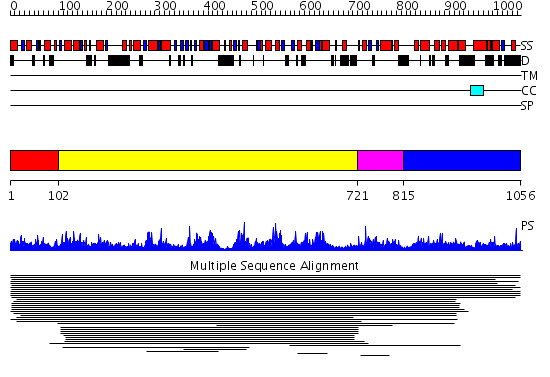
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [102..720] | 4.027892 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [721..814] | 1.020986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [815..1056] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)