













| General Information: |
|
| Name(s) found: |
HHT2 /
YNL031C
[SGD]
HHT1 / YBR010W [SGD] |
| Description(s) found:
Found 60 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 136 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear nucleosome [TAS |
| Biological Process: |
chromatin assembly or disassembly
[TAS |
| Molecular Function: |
DNA binding
[TAS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
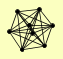
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ASF1 HAT1 HAT2 HHF1, HHF2 HHT1, HHT2 HIF1 POB3 PSH1 |
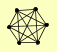
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
HHF1, HHF2 HHT1, HHT2 HTA1 HTA2 HTB1 HTB2 |
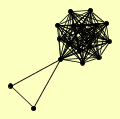
| View Details | Krogan NJ, et al. (2006) |
|
ARA2 AYT1 BRO1 CDC73 CTR9 GLO4 HHT1, HHT2 LEO1 PAF1 PML39 POB3 PSH1 RTF1 SEN1 SKO1 SPT16 VAC7 YHR035W |
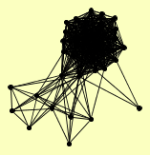
| View Details | Ho Y, et al. (2002) |
|
AHP1 AMS1 BIK1 CKA1 CKB1 CKB2 CPR1 DBP2 DUN1 EGD1 ENP2 ERB1 FPR3 HAS1 HHT1, HHT2 HOT1 HTA1 HTB1 HTB2 KRE33 KRI1 LAP4 MAG1 MGM101 MGT1 NOG1 NOP12 NOP2 NOP4 NOP56 NUG1 PDI1 POB3 PUF6 PWP1 SPT16 SSF1 TIF6 VMA4 YER084W |
| View Details | Qiu et al. (2008) |
|
HHF1, HHF2 HHT1, HHT2 HTA1 HTA2 HTB1 HTB2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
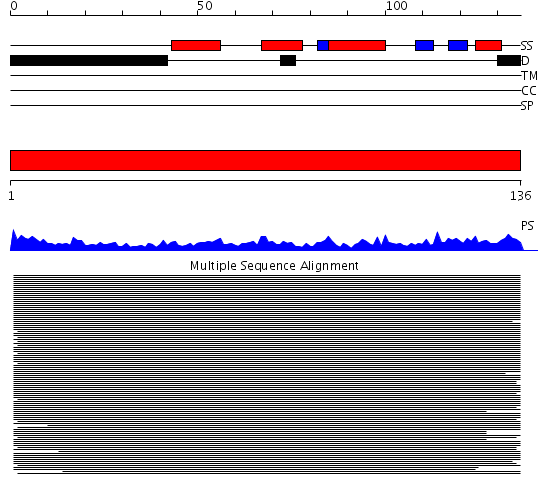
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | 542.626727 | Histone H3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)