













| General Information: |
|
| Name(s) found: |
VAC7 /
YNL054W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1165 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA integral to membrane [IDA fungal-type vacuole [IDA |
| Biological Process: |
vacuole inheritance
[IMP phospholipid metabolic process [IGI |
| Molecular Function: |
enzyme activator activity
[IMP enzyme regulator activity [IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEEDRKLTV ETETVEAPVA NNLLLSNNSN VVAPNPSIPS ASTSTSPLHR EIVDDSVATA 60
61 NTTSNVVQHN LPTIDNNLMD SDATSHNQDH WHSDINRAGT SMSTSDIPTD LHLEHIGSVS 120
121 STNNNSNNAL INHNPLSSHL SNPSSSLRNK KSSLLVASNP AFASDVELSK KKPAVISNNM 180
181 PTSNIALYQT ARSANIHGPS STSASKAFRK ASAFSNNTAP STSNNIGSNT PPAPLLPLPS 240
241 LSQQNKPKII ERPTMHVTNS REILLGENLL DDTKAKNAPA NSTTHDNGPV ANDGLRIPNH 300
301 SNADDNENNN KMKKNKNINS GKNERNDDTS KICTTSTKTA PSTAPLGSTD NTQALTASVS 360
361 SSNADNHNNN KKKTSSNNNG NNSNSASNKT NADIKNSNAD LSASTSNNNA INDDSHESNS 420
421 EKPTKADFFA ARLATAVGEN EISDSEETFV YESAANSTKN LIFPDSSSQQ QQQQQQPPKQ 480
481 QQQQQNHGIT SKISAPLLNN NKKLLSRLKN SRHISTGAIL NNTIATISTN PNLNSNVMQN 540
541 NNNLMSGHNH LDELSSIKQE PPHQLQQQQP PMDVQSVDSY TSDNPDSNVI AKSPDKRSSL 600
601 VSLSKVSPHL LSSTSSNGNT ISCPNVATNS QELEPNNDIS TKKSLSNSTL RHSSANRNSN 660
661 YGDNKRPLRT TVSKIFDSNP NGAPLRRYSG VPDHVNLEDY IEQPHNYPTM QNSVKKDEFY 720
721 NSRNNKFPHG LNFYGDNNVI EEENNGDSSN VNRPQHTNLQ HEFIPEDNES DENDIHSMFY 780
781 YNHKNDLETK PLISDYGEDE DVDDYDRPNA TFNSYYGSAS NTHELPLHGR MPSRSNNDYY 840
841 DFMVGNNTGN NNQLNEYTPL RMKRGQRHLS RTNNSIMNGS IHMNGNDDVT HSNINNNDIV 900
901 GYSPHNFYSR KSPFVKVKNF LYLAFVISSL LMTGFILGFL LATNKELQDV DVVVMDNVIS 960
961 SSDELIFDIT VSAFNPGFFS ISVSQVDLDI FAKSSYLKCD SNGDCTVMEQ ERKILQITTN1020
1021 LSLVEESANN DISGGNIETV LLGTAKKLET PLKFQGGAFN RNYDVSVSSV KLLSPGSREA1080
1081 KHENDDDDDD DGDDGDDENN TNERQYKSKP NARDDKEDDT KKWKLLIKHD YELIVRGSMK1140
1141 YEVPFFNTQK STAIQKDSMV HPGKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
HHT1, HHT2 SKO1 VAC7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
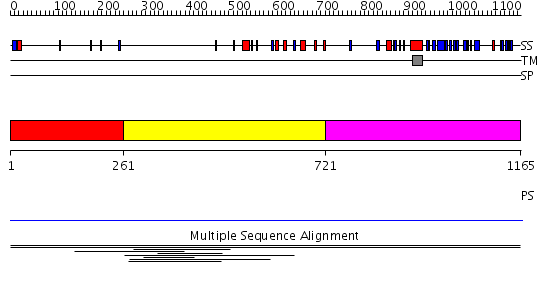
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..260] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [261..720] | 1.005997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [721..1165] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [758..905] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [906..1165] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|