













| General Information: |
|
| Name(s) found: |
VAM3 /
YOR106W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 283 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar membrane
[IDA |
| Biological Process: |
vesicle fusion
[TAS Golgi to vacuole transport [TAS |
| Molecular Function: |
receptor signaling protein serine/threonine kinase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFFDIEAQS SKGNSQQEPQ FSTNQKTKEL SNLIETFAEQ SRVLEKECTK IGSKRDSKEL 60
61 RYKIETELIP NCTSVRDKIE SNILIHQNGK LSADFKNLKT KYQSLQQSYN QRKSLFPLKT 120
121 PISPGTSKER KDIHPRTEAV RQDPESSYIS IKVNEQSPLL HNEGQHQLQL QEEQEQQQQG 180
181 LSQEELDFQT IIHQERSQQI GRIHTAVQEV NAIFHQLGSL VKEQGEQVTT IDENISHLHD 240
241 NMQNANKQLT RADQHQRDRN KCGKVTLIII IVVCMVVLLA VLS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
VAM3 VAM7 |
| View Details | Qiu et al. (2008) |
|
PEP12 SED5 SSO1 VAM3 VMA9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample: hx1 | Hao Xu, et al. (2010) | |
| View Run | sample: hx2 | Hao Xu, et al. (2010) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | sample: hx4 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx21: both fusion and trans-SNARE complex formation take place. Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
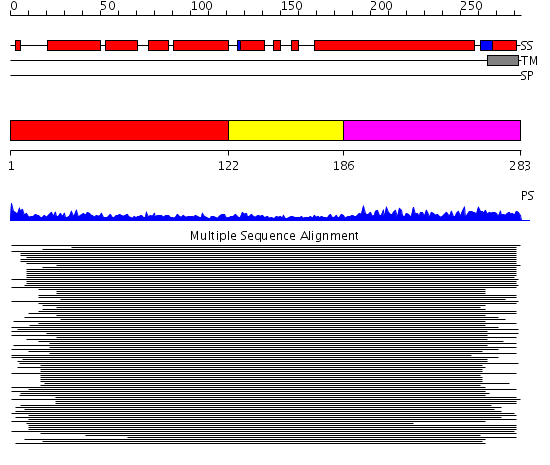
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 395.30103 | Vam3p N-terminal domain | |
| 2 | View Details | [122..185] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [186..283] | 13.98 | Syntaxin 7 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|