













| General Information: |
|
| Name(s) found: |
MLP2 /
YIL149C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1679 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA ribonucleoprotein complex [IPI mitochondrion [IDA nucleoplasm [IDA |
| Biological Process: |
mRNA export from nucleus
[IGI protein import into nucleus [IGI regulation of transcription [IEP |
| Molecular Function: |
ribonucleoprotein binding
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDKISEFLN VPFESLQGVT YPVLRKLYKK IAKFERSEEE VTKLNVLVDE IKSQYYSRIS 60
61 KLKQLLDESS EQKNTAKEEL NGLKDQLNEE RSRYRREIDA LKKQLHVSHE AMREVNDEKR 120
121 VKEEYDIWQS RDQGNDSLND DLNKENKLLR RKLMEMENIL QRCKSNAISL QLKYDTSVQE 180
181 KELMLQSKKL IEEKLSSFSK KTLTEEVTKS SHVENLEEKL YQMQSNYESV FTYNKFLLNQ 240
241 NKQLSQSVEE KVLEMKNLKD TASVEKAEFS KEMTLQKNMN DLLRSQLTSL EKDCSLRAIE 300
301 KNDDNSCRNP EHTDVIDELI DTKLRLEKSK NECQRLQNIV MDCTKEEEAT MTTSAVSPTV 360
361 GKLFSDIKVL KRQLIKERNQ KFQLQNQLED FILELEHKTP ELISFKERTK SLEHELKRST 420
421 ELLETVSLTK RKQEREITSL RQKINGCEAN IHSLVKQRLD LARQVKLLLL NTSAIQETAS 480
481 PLSQDELISL RKILESSNIV NENDSQAIIT ERLVEFSNVN ELQEKNVELL NCIRILADKL 540
541 ENYEGKQDKT LQKVENQTIK EAKDAIIELE NINAKMETRI NILLRERDSY KLLASTEENK 600
601 ANTNSVTSME AAREKKIREL EAELSSTKVE NSAIIQNLRK ELLIYKKSQC KKKTTLEDFE 660
661 NFKGLAKEKE RMLEEAIDHL KAELEKQKSW VPSYIHVEKE RASTELSQSR IKIKSLEYEI 720
721 SKLKKETASF IPTKESLTRD FEQCCKEKKE LQMRLKESEI SHNENKMDFS SKEGQYKAKI 780
781 KELENNLERL RSDLQSKIQE IESIRSCKDS QLKWAQNTID DTEMKMKSLL TELSNKETTI 840
841 EKLSSEIENL DKELRKTKFQ YKFLDQNSDA STLEPTLRKE LEQIQVQLKD ANSQIQAYEE 900
901 IISSNENALI ELKNELAKTK ENYDAKIELE KKEKWAREED LSRLRGELGE IRALQPKLKE 960
961 GALHFVQQSE KLRNEVERIQ KMIEKIEKMS TIVQLCKKKE MSQYQSTMKE NKDLSELVIR1020
1021 LEKDAADCQA ELTKTKSSLY SAQDLLDKHE RKWMEEKADY ERELISNIEQ TESLRVENSV1080
1081 LIEKVDDTAA NNGDKDHLKL VSLFSNLRHE RNSLETKLTT CKRELAFVKQ KNDSLEKTIN1140
1141 DLQRTQTLSE KEYQCSAVII DEFKDITKEV TQVNILKENN AILQKSLKNV TEKNREIYKQ1200
1201 LNDRQEEISR LQRDLIQTKE QVSINSNKIL VYESEMEQCK QRYQDLSQQQ KDAQKKDIEK1260
1261 LTNEISDLKG KLSSAENANA DLENKFNRLK KQAHEKLDAS KKQQAALTNE LNELKAIKDK1320
1321 LEQDLHFENA KVIDLDTKLK AHELQSEDVS RDHEKDTYRT LMEEIESLKK ELQIFKTANS1380
1381 SSDAFEKLKV NMEKEKDRII DERTKEFEKK LQETLNKSTS SEAEYSKDIE TLKKEWLKEY1440
1441 EDETLRRIKE AEENLKKRIR LPSEERIQKI ISKRKEELEE EFRKKLKENA GSLTFLDNKG1500
1501 SGEDAEEELW NSPSKGNSER PSAVAGFINQ KNLKPQEQLK NVKNDVSFND SQSMVTNKEN1560
1561 NIVDSSAAGN KAIPTFSFGK PFFSSNTSSL QSFQNPFTAS QSNINTNAPL RTLNIQPEVA1620
1621 VKAAINFSNV TDLTNNSTDG AKITEIGSTS KRPIESGTSS DPDTKKVKES PANDQASNE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #33 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) | |
| View Run | #22 Asynchronous Prep2-IMAC Phosphopeptide enrichment B1-2 | Keck JM, et al. (2011) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #35 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #01 Alpha factor Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #28 Asynchronous Prep5-TiO2 Phosphopeptide Enrichment | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #16 Mitotic-New search criteria | Keck JM, et al. (2011) | |
| View Run | #15 Mitotic 2nd-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #09 Alpha Factor Prep4-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #18 Mitotic Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
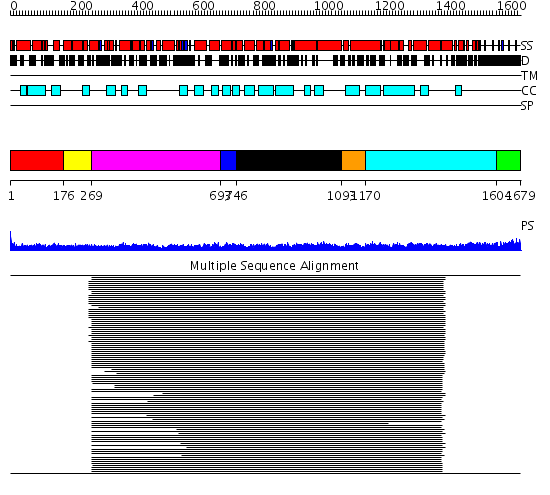
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..175] | 1.465995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [176..268] | 9.522879 | Tropomyosin | |
| 3 | View Details | [269..692] | 48.980454 | Heavy meromyosin subfragment | |
| 4 | View Details | [693..745] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [746..1092] | 18.998997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1093..1169] | N/A | Confident ab initio structure predictions are available. | |
| 7 | View Details | [1170..1603] | 16.682998 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1604..1679] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)