













| General Information: |
|
| Name(s) found: |
ADE3 /
YGR204W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 946 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
folic acid and derivative metabolic process
[TAS]
purine base biosynthetic process [IMP |
| Molecular Function: |
formate-tetrahydrofolate ligase activity
[IMP methenyltetrahydrofolate cyclohydrolase activity [IMP methylenetetrahydrofolate dehydrogenase (NADP+) activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGQVLDGKA CAQQFRSNIA NEIKSIQGHV PGFAPNLAII QVGNRPDSAT YVRMKRKAAE 60
61 EAGIVANFIH LDESATEFEV LRYVDQLNED PHTHGIIVQL PLPAHLDEDR ITSRVLAEKD 120
121 VDGFGPTNIG ELNKKNGHPF FLPCTPKGII ELLHKANVTI EGSRSVVIGR SDIVGSPVAE 180
181 LLKSLNSTVT ITHSKTRDIA SYLHDADIVV VAIGQPEFVK GEWFKPRDGT SSDKKTVVID 240
241 VGTNYVADPS KKSGFKCVGD VEFNEAIKYV HLITPVPGGV GPMTVAMLMQ NTLIAAKRQM 300
301 EESSKPLQIP PLPLKLLTPV PSDIDISRAQ QPKLINQLAQ ELGIYSHELE LYGHYKAKIS 360
361 PKVIERLQTR QNGKYILVSG ITPTPLGEGK STTTMGLVQA LTAHLGKPAI ANVRQPSLGP 420
421 TLGVKGGAAG GGYSQVIPMD EFNLHLTGDI HAIGAANNLL AAAIDTRMFH ETTQKNDATF 480
481 YNRLVPRKNG KRKFTPSMQR RLNRLGIQKT NPDDLTPEEI NKFARLNIDP DTITIKRVVD 540
541 INDRMLRQIT IGQAPTEKNH TRVTGFDITV ASELMAILAL SKDLRDMKER IGRVVVAADV 600
601 NRSPVTVEDV GCTGALTALL RDAIKPNLMQ TLEGTPVLVH AGPFANISIG ASSVIADRVA 660
661 LKLVGTEPEA KTEAGYVVTE AGFDFTMGGE RFFNIKCRSS GLTPNAVVLV ATVRALKSHG 720
721 GAPDVKPGQP LPSAYTEENI EFVEKGAANM CKQIANIKQF GVPVVVAINK FETDTEGEIA 780
781 AIRKAALEAG AFEAVTSNHW AEGGKGAIDL AKAVIEASNQ PVDFHFLYDV NSSVEDKLTT 840
841 IVQKMYGGAA IDILPEAQRK IDMYKEQGFG NLPICIAKTQ YSLSHDATLK GVPTGFTFPI 900
901 RDVRLSNGAG YLYALAAEIQ TIPGLATYAG YMAVEVDDDG EIDGLF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ADE3 MIS1 RPP1B TMA108 |
| View Details | Gavin AC, et al. (2006) |
|
ADE3 CSE1 DPS1 SAR1 TDH1 TOM1 YDR341C YNK1 |
| View Details | Gavin AC, et al. (2002) |
|
ADE3 ARX1 DBP2 LSG1 MIS1 NOG1 NOP7 NUG1 RAT1 SHM1 YPL009C |
| View Details | Ho Y, et al. (2002) |
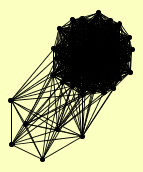
|
ACS2 ADE3 ADE6 ADO1 ALA1 APE2 ARA1 BAT1 CDC10 CLU1 CYS3 DED81 DPS1 ERG10 ERG20 FPR1 GCY1 GPH1 GRS1 HOM2 HOM3 HYP2 ILS1 IMD3 KRS1 LSP1 MDH3 MLP2 OLA1 PAB1 PFK2 PGM2 PMI40 PRP6 SAC6 SCP160 SOD1 SSZ1 THR4 THS1 TRR1 UBA1 URA1 YDL124W YDR341C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
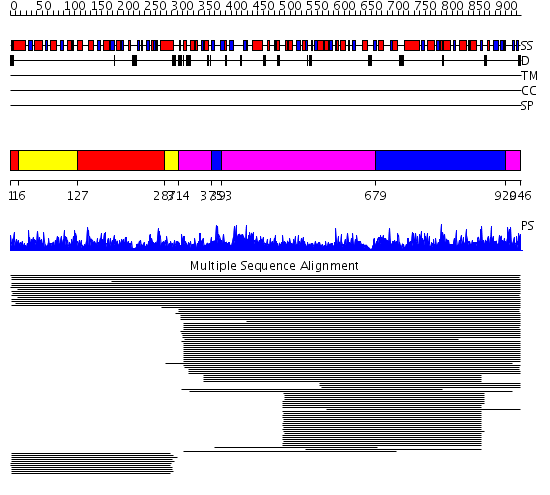
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..15] [127..286] |
848.9897 | Methylenetetrahydrofolate dehydrogenase/cyclohydrolase; Tetrahydrofolate dehydrogenase/cyclohydrolase | |
| 2 | View Details | [16..126] [287..313] |
848.9897 | Methylenetetrahydrofolate dehydrogenase/cyclohydrolase; Tetrahydrofolate dehydrogenase/cyclohydrolase | |
| 3 | View Details | [314..374] [393..678] [920..946] |
2400.0 | Formyltetrahydrofolate synthetase | |
| 4 | View Details | [375..392] [679..919] |
2400.0 | Formyltetrahydrofolate synthetase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)