













| General Information: |
|
| Name(s) found: |
GPH1 /
YPR160W
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 902 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IC |
| Biological Process: |
glycogen catabolic process
[IMP |
| Molecular Function: |
glycogen phosphorylase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPASTSTTN DMITEEPTSP HQIPRLTRRL TGFLPQEIKS IDTMIPLKSR ALWNKHQVKK 60
61 FNKAEDFQDR FIDHVETTLA RSLYNCDDMA AYEAASMSIR DNLVIDWNKT QQKFTTRDPK 120
121 RVYYLSLEFL MGRALDNALI NMKIEDPEDP AASKGKPREM IKGALDDLGF KLEDVLDQEP 180
181 DAGLGNGGLG RLAACFVDSM ATEGIPAWGY GLRYEYGIFA QKIIDGYQVE TPDYWLNSGN 240
241 PWEIERNEVQ IPVTFYGYVD RPEGGKTTLS ASQWIGGERV LAVAYDFPVP GFKTSNVNNL 300
301 RLWQARPTTE FDFAKFNNGD YKNSVAQQQR AESITAVLYP NDNFAQGKEL RLKQQYFWCA 360
361 ASLHDILRRF KKSKRPWTEF PDQVAIQLND THPTLAIVEL QRVLVDLEKL DWHEAWDIVT 420
421 KTFAYTNHTV MQEALEKWPV GLFGHLLPRH LEIIYDINWF FLQDVAKKFP KDVDLLSRIS 480
481 IIEENSPERQ IRMAFLAIVG SHKVNGVAEL HSELIKTTIF KDFVKFYGPS KFVNVTNGIT 540
541 PRRWLKQANP SLAKLISETL NDPTEEYLLD MAKLTQLGKY VEDKEFLKKW NQVKLNNKIR 600
601 LVDLIKKEND GVDIINREYL DDTLFDMQVK RIHEYKRQQL NVFGIIYRYL AMKNMLKNGA 660
661 SIEEVAKKYP RKVSIFGGKS APGYYMAKLI IKLINCVADI VNNDESIEHL LKVVFVADYN 720
721 VSKAEIIIPA SDLSEHISTA GTEASGTSNM KFVMNGGLII GTVDGANVEI TREIGEDNVF 780
781 LFGNLSENVE ELRYNHQYHP QDLPSSLDSV LSYIESGQFS PENPNEFKPL VDSIKYHGDY 840
841 YLVSDDFESY LATHELVDQE FHNQRSEWLK KSVLSVANVG FFSSDRCIEE YSDTIWNVEP 900
901 VT |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
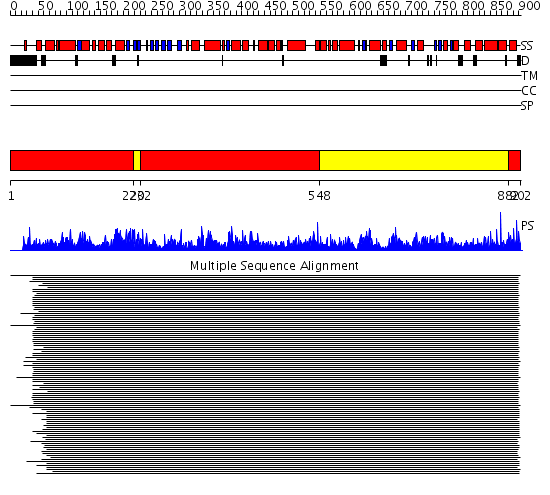
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..219] [232..547] [882..902] |
11000.0 | Glycogen phosphorylase | |
| 2 | View Details | [220..231] [548..881] |
11000.0 | Glycogen phosphorylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)