













| General Information: |
|
| Name(s) found: |
FIN1 /
YDR130C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 291 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spindle pole body [IDA spindle [IDA intermediate filament [IDA astral microtubule [IDA |
| Biological Process: |
mitotic sister chromatid segregation
[IMP intermediate filament-based process [IDA mitotic spindle stabilization [IMP |
| Molecular Function: |
structural constituent of cytoskeleton
[IDA microtubule binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNKSNRRSL RDIGNTIGRN NIPSDKDNVF VRLSMSPLRT TSQKEFLKPP MRISPNKTDG 60
61 MKHSIQVTPR RIMSPECLKG YVSKETQSLD RPQFKNSNKN VKIQNSDHIT NIIFPTSPTK 120
121 LTFSNENKIG GDGSLTRIRA RFKNGLMSPE RIQQQQQQHI LPSDAKSNTD LCSNTELKDA 180
181 PFENDLPRAK LKGKNLLVEL KKEEEDVGNG IESLTKSNTK LNSMLANEGK IHKASFQKSV 240
241 KFKLPDNIVT EETVELKEIK DLLLQMLRRQ REIESRLSNI ELQLTEIPKH K |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
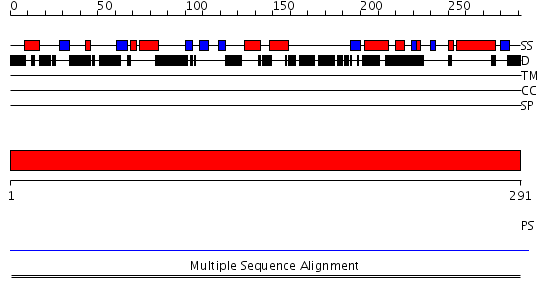
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..291] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)