













| General Information: |
|
| Name(s) found: |
ZDS1 /
YMR273C
[SGD]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 915 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud tip
[IDA cytoplasm [IPI cellular bud neck [IDA nuclear pore [IPI |
| Biological Process: |
mRNA export from nucleus
[IGI chromatin silencing [IMP cell aging [IMP |
| Molecular Function: |
protein binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNRDNESML RTTSSDKAIA SQRDKRKSEV LIAAQSLDNE IRSVKNLKRL SIGSMDLLID 60
61 PELDIKFGGE SSGRRSWSGT TSSSASMPSD TTTVNNTRYS DPTPLENLHG RGNSGIESSN 120
121 KTKQGNYLGI KKGVHSPSRK LNANVLKKNL LWVPANQHPN VKPDNFLELV QDTLQNIQLS 180
181 DNGEDNDGNS NENNDIEDNG EDKESQSYEN KENNTINLNR GLSRHGNASL IRRPSTLRRS 240
241 YTEFDDNEDD DNKGDSASET VNKVEERISK IKERPVSLRD ITEELTKISN SAGLTDNDAI 300
301 TLARTLSMAG SYSDKKDQPQ PEGHYDEGDI GFSTSQANTL DDGEFASNMP INNTMTWPER 360
361 SSLRRSRFNT YRIRSQEQEK EVEQSVDEMK NDDEERLKLT KNTIKVEIDP HKSPFRQQDE 420
421 DSENMSSPGS IGDFQDIYNH YRQSSGEWEQ EMGIEKEAEE VPVKVRNDTV EQDLELREGT 480
481 TDMVKPSATD DNKETKRHRR RNGWTWLNNK MSREDDNEEN QGDDENEENV DSQRMELDNS 540
541 KKHYISLFNG GEKTEVSNKE EMNNSSTSTA TSQTRQKIEK TFANLFRRKP HHKHDASSSP 600
601 SSSPSSSPSI PNNDAVHVRV RKSKKLGNKS GREPVEPIVL RNRPRPHRHH HSRHGSQKIS 660
661 VKTLKDSQPQ QQIPLQPQLE GAIEIEKKEE SDSESLPQLQ PAVSVSSTKS NSRDREEEEA 720
721 KKKNKKRSNT TEISNQQHSK HVQKENTDEQ KAQLQAPAQE QVQTSVPVQA SAPVQNSAPV 780
781 QTSAPVEASA QTQAPAAPPL KHTSILPPRK LTFADVKKPD KPNSPVQFTD SAFGFPLPLL 840
841 TVSTVIMFDH RLPINVERAI YRLSHLKLSN SKRGLREQVL LSNFMYAYLN LVNHTLYMEQ 900
901 VAHDKEQQQQ QQQQP |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
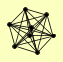
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CDC55 PPH21 PPH22 RRD2 RTS1 TPD3 ZDS1 ZDS2 |
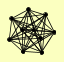
| View Details | Krogan NJ, et al. (2006) |
|
CDC55 FIR1 PHR1 PPH21 PPH22 RRD2 TPD3 YRR1 ZDS1 ZDS2 |
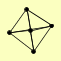
| View Details | Gavin AC, et al. (2006) |
|
CDC55 RTS3 TPD3 ZDS1 ZDS2 |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC55 CIN1 ERG13 GUS1 HHF1, HHF2 HOS2 HOS4 IML1 KAP95 KEL1 LTE1 MYO5 PFK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RRD2 RTS1 RTS3 SCL1 SET3 SIF2 SNT1 SRP1 TDH2 TDH3 TEF4 TPD3 YBL104C YOR1 YRA1 ZDS1 ZDS2 |
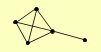
| View Details | Qiu et al. (2008) |
|
BEM1 BOI1 BOI2 SUS1 ZDS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample zds1 from august 2005 | McCusker D, et al (2007) | |
| View Run | Sample zds1 gel band from august 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Drees BL, et al. (2001) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
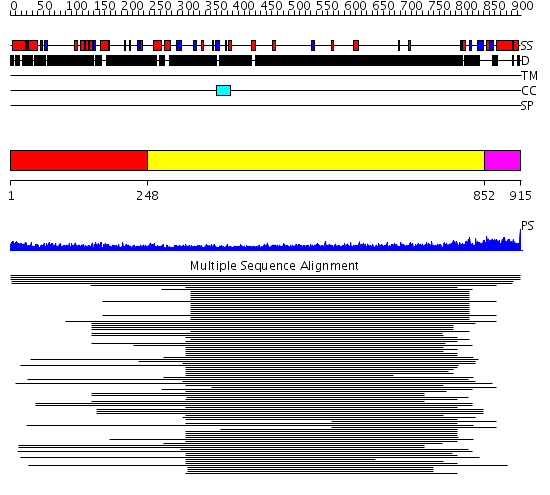
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..247] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [248..851] | 30.269998 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [852..915] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [460..605] | 8.0 | Solution Structure of Choline Binding Protein A, Domain R2, the Major Adhesin of Streptococcus pneumoniae | |
| 5 | View Details | [606..774] | 3.329996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [775..837] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [838..915] | 1.057994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.74 |
Source: Reynolds et al. (2008)