













| General Information: |
|
| Name(s) found: |
LTE1 /
YAL024C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1435 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud
[TAS |
| Biological Process: |
vesicle-mediated transport
[IGI regulation of exit from mitosis [TAS response to drug [IMP mitotic cell cycle spindle orientation checkpoint [TAS |
| Molecular Function: |
guanyl-nucleotide exchange factor activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEIFSQKDYY PTPSSNVISY ESDCVSKPVN SADLPALIVH LSSPLEGVDY NASADFFLIY 60
61 RNFITPQDLH DLLIYRFRWC IREITTNAAK AKRRRIGEVA LVRTFVLLRH SILNYFVQDF 120
121 LPNITLRLRL IEFLNDKHIE QYPKIISSCI INLKKNWVHC SKLVWENIEL NEPDKLDFDA 180
181 WLHYSLKDFT QLESLHKRGS RLSIYARQSF ASPDFRNQSV LSLYKTSDVF RLPEKLQSSN 240
241 SSKNQRSPSM LLFPDNTSNV YSKHRIAKEP SVDNESEDMS DSKQKISHLS KVTLVSTLMK 300
301 GVDYPSSYAV DKIMPPTPAK KVEFILNSLY IPEDLNEQSG TLQGTSTTSS LDNNSNSNSR 360
361 SNTSSMSVLH RSAIGLLAKW MKNHNRHDSS NDKKFMSAIK PANQKPEMDA FVKYVVSISS 420
421 LNRKSSKEEE EEFLNSDSSK FDILSARTID EVESLLHLQN QLIEKVQTHS NNNRGPTVNV 480
481 DCERREHIHD IKILQQNSFK PSNDNFSAMD NLDLYQTVSS IAQSVISLTN TLNKQLQNNE 540
541 SNMQPSPSYD ALQRRKVKSL TTAYYNKMHG SYSAESMRLF DKDNSSSRTD ENGPQRLLFH 600
601 ETDKTNSEAI TNMTPRRKNH SQSQKSMTSS PLKNVLPDLK ESSPLNDSRE DTESITYSYD 660
661 SELSSSSPPR DTVTKKSRKV RNIVNNTDSP TLKTKTGFLN LREFTFEDTK SLDEKKSTID 720
721 GLEKNYDNKE NQESEYESTK KLDNSLDASS EANNYDITTR KKHSSCNHKI KQAVVRPASG 780
781 RISISRVQSI AITPTKELSI VDPEQNKSNS VIEEISEIEP LNLEYNKKSA LYSDTSSTVI 840
841 SISTSKLFES AQNSPLKQTQ NPQREFPNGT SVSETNRIRL SIAPTIESVV SDLNSITTGS 900
901 TVETFETSRD LPVPHQRIIN LREEYQRGNQ DIISNTSSLH ELKTIDLSDS NNDLESPSTH 960
961 AKNNKYFFSP DDGSIDVASP MKNVEELKSK FLKNESETNS NISGSVLTMD DIDINDTSSA1020
1021 RNTRRANSES AFTGSLNKKN LNEIANMLDD SINDDPITVA LMKLEGTYEK IPEKPENTKS1080
1081 SDAIGIKTSK LADEVEMLNL NNLPSFQNSP AEKRKSLLIE RRRQTIMNIP FTPDQSEKEG1140
1141 FTSSSPEKID VSANVDVAVQ AAQIQELIGQ YRIHDSRLMI SNNESHVPFI LMYDSLSVAQ1200
1201 QMTLIEKEIL GEIDWKDLLD LKMKHEGPQV ISWLQLLVRN ETLSGIDLAI SRFNLTVDWI1260
1261 ISEILLTKSS KMKRNVIQRF IHVADHCRTF QNFNTLMEII LALSSSVVQK FTDAWRLIEP1320
1321 GDLLTWEELK KIPSLDRNYS TIRNLLNSVN PLVGCVPFIV VYLSDLSANA EKKDWILEDK1380
1381 VVNYNKFDTN VQIVKNFIQR VQWSKFYTFK VNHELLSKCV YISTLTQEEI NELST |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
LTE1 RPS3 |
| View Details | Krogan NJ, et al. (2006) |
|
AI1 LTE1 TAZ1 |
| View Details | Gavin AC, et al. (2006) |
|
BEM2 KEL1 LTE1 |
| View Details | Gavin AC, et al. (2002) |

|
BLM10 CDC55 CIN1 ERG13 GUS1 HHF1, HHF2 HOS2 HOS4 IML1 KAP95 KEL1 LTE1 MYO5 PFK1 PPH21 PPH22 PRE1 PRE10 PRE2 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RRD2 RTS1 RTS3 SCL1 SET3 SIF2 SNT1 SRP1 TDH2 TDH3 TEF4 TPD3 YBL104C YOR1 YRA1 ZDS1 ZDS2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
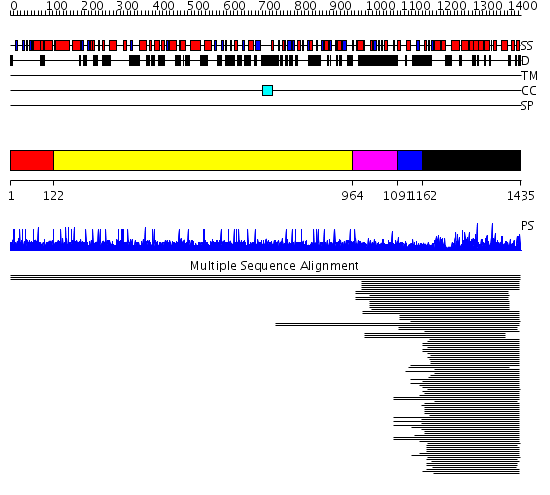
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 23.154902 | Guanine nucleotide exchange factor for Ras-like GTPases; N-terminal motif No confident structure predictions are available. | |
| 2 | View Details | [122..963] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [964..1090] | 1.012983 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1091..1161] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1162..1435] | 109.457575 | Son of sevenless protein homolog 1 (sos-1) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)