













| General Information: |
|
| Name(s) found: |
BEM2 /
YER155C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2167 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA intracellular [TAS |
| Biological Process: |
small GTPase mediated signal transduction
[IPI cellular cell wall organization [IPI microtubule cytoskeleton organization [IGI budding cell bud growth [IPI actin cytoskeleton organization [IGI |
| Molecular Function: |
Rho GTPase activator activity
[IDA signal transducer activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKGLLWSKNR KSSTASASSS STSTSHKTTT ASTASSSSPS SSSQTIRNST SGASPYMHSH 60
61 HHHGQGHSHH RGEDNNRDKR KSSVFPPSKQ YTSTSSSQVN LGMYHSDTNT RSSRSIASTL 120
121 KDDSPSVCSE DEISNSSSQK SNAQDETPIA YKKSAHSKDS LLPSRSSSLS PPQSRCSTGT 180
181 TLEKSLNTSG ISNSSGTNNN NSNNNNDNEQ KQRNVIHLNS ENYDTTVFKT GWVNKSHGQT 240
241 VATNYNSSMT APSSSSSSSS QNLRNDAYSR NRESRFYGND GSSLKNDDSS STTATNSGND 300
301 VASARSSMAI DPQMLVPDYR LYRAQLKGCV LNLYKSGLNS NIKFFDPTLP ASNSSIANEN 360
361 HQQKKQQTNN QAQAEALHQK QSFGQMGEPI TLDLKYLSEV YPHPDLRQDS DGKIISGTIE 420
421 SLCHTVLFYP GPKQSDVPNE KSLSKTHRAV INLLLMFPLL DHFIKFLKVF NQFGLSFTKN 480
481 KSRLTNNSTQ FYNISPAVDD SMTQRLALTA KTILDVFPGF LLDEPMLKTI ISLLDTISLH 540
541 NDEISNNLKI KIANKHNELM KLTAFTRSLP MATSSTHELE IILDPSHFLS LDITTLADEV 600
601 HHINLKFDKV WAPKFDYSLL YDSKFINRRI VSLNPLVFNN DQNIHFLGRL LISHLFPTNP 660
661 EFSKKVTPKV RAELLDKWVQ IGCRFEHLGD MVSWLAVATI ICSIPVLRSS SWKYVPDQSL 720
721 KTIFKDWVPT IIQLERRQRT SKSTSSVFIL APPNLDDDFT RANVISYFGD LLIHADDLPS 780
781 DTKFKYLEKK INRTKNAFHK WQQRLQAIDS TRHKTNSTEN VRDNDSPNNV VYQLWKFHLS 840
841 QPPLNIEGIM KLSVQHEPPI IDQKAYSTIG SQRSALVTGS YLPILFNELF PNYSLFPKNT 900
901 LVGAASDAKL PPPRSSARLS KSLSISEPIP IASNSHTMGS LTDDAMSSKN DNNKVTGVGK 960
961 IDGPVIKEMS SKQSNKQRLL KSVRDVFNID MDVFHISDEL VFKSVYDNDG KSRPASMVIE1020
1021 TPKRFSQHSS MLINNPATPN QKMRDSLDTT GRLSKTLENM DFFNNIGQVS DSLKESIIRV1080
1081 VLKSSSLEKI FDLLVLTSNI FSKLVDTKDL ENYYYHQRQR GHSTRGLSDD NIGLLDYAFV1140
1141 KLTMDNDIFT ETFFNTYKSF TTTTTVLENM AKRYVGAKSC SVSISKILDR SDDSKMKINE1200
1201 DTNLVSSSLY DQNFPVWDMK VTDDENINLI YMAKIQIGAA EAILHLVKNH YSDFTDDLCN1260
1261 NSTLLDIIKI MEQEVSTEWP TRIANSKLQK SLPENFVIET ENLLTTLTDL FHGIKSAYQK1320
1321 QLYRPIGVNR TQKRITDILN SFNTFSFTDL NNIIDDPSFS DDMIRSFQKL HSTNYEDILE1380
1381 WIYQLDNFIS KKFNLVSKKD WIVLFQELEL LSKESLVSFF NYPLHFKSSK LINPGYLQLH1440
1441 EFEISNLFTW ISTLILKDDN GTESLFFEKL PQSIKLLIKL HTSLTTFFVM EISNVNKSSS1500
1501 ERLTTCKVIL QILNYIRWKN GSLDLFDSEE DESPHAICPH IPAFIETAIA HAIISPESRN1560
1561 YELSWIKASE KLSDPTKGTQ NLRSISNVLE KIDDIHIKRF IEIDDVFSKN CKNLCPCPGW1620
1621 FISRLLEISQ FVPNMSITNS KLINFDKRRF VNNIISNVLD LIPNEREFPL DIEMSDENPS1680
1681 KRTTFGRILF NNFEDVNKVY RKKTKKVSES EAISERFQEQ GVFNEILVNE IEKIKREARK1740
1741 LEVLLDQEKI LKNSAALHQA VPKKNRKSVI ISGTHSDNDH SYNINKNTGQ TPSLGSVMES1800
1801 NNSARNRRDS RASFSTNRSS VVSNSSHNGV SKKIGGFFRR PFSIGGFNTS SSNYSLNSIL1860
1861 SQEVSSNKSI LPSILPEVDS MQLHDLKPSY SLKTFEIKSI MEIINHRNIP AYYYAFKIVM1920
1921 QNGHEYLIQT ASSSDLTEWI KMIKASKRFS FHSKKYKGKT HNKIFGVPLE DVCERENTLI1980
1981 PTIVVKLLEE IELRGLDEVG LYRIPGSIGS INALKNAFDE EGATDNSFTL EDDRWFEVNA2040
2041 IAGCFKMYLR ELPDSLFSHA MVNDFTDLAI KYKAHAMVNE EYKRMMNELL QKLPTCYYQT2100
2101 LKRIVFHLNK VHQHVVNNKM DASNLAIVFS MSFINQEDLA NSMGSRLGAV QTILQDFIKN2160
2161 PNDYFKQ |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
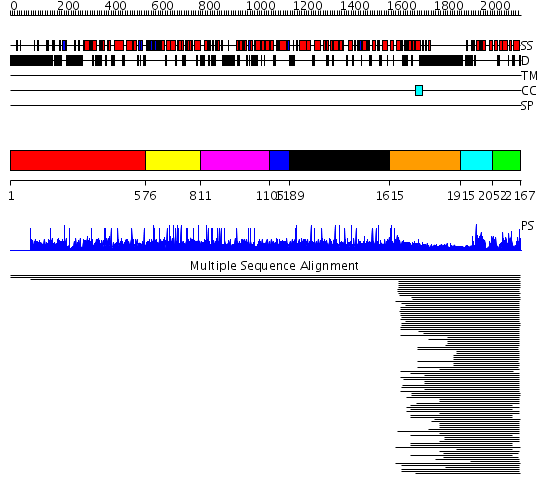
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..575] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [576..810] | 95.744727 | RasGEF domain No confident structure predictions are available. | |
| 3 | View Details | [811..1104] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1105..1188] | 22.853872 | Guanine nucleotide exchange factor for Ras-like GTPases; N-terminal motif No confident structure predictions are available. | |
| 5 | View Details | [1189..1614] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1615..1914] | 2.146989 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1915..2051] | 124.211549 | Graf | |
| 8 | View Details | [2052..2167] | 124.211549 | Graf | |
| 9 | View Details | [1873..2167] | 71.30103 | Crystal Structure of the Human Beta2-Chimaerin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|