













| General Information: |
|
| Name(s) found: |
EHD3 /
YDR036C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 500 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
[NOT]fatty acid beta-oxidation
[IMP endocytosis [IMP |
| Molecular Function: |
3-hydroxyisobutyryl-CoA hydrolase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRNTLKCAQ LSSKYGFKTT TRTFMTTQPQ LNVTDAPPVL FTVQDTARVI TLNRPKKLNA 60
61 LNAEMSESMF KTLNEYAKSD TTNLVILKSS NRPRSFCAGG DVATVAIFNF NKEFAKSIKF 120
121 FTDEYSLNFQ IATYLKPIVT FMDGITMGGG VGLSIHTPFR IATENTKWAM PEMDIGFFPD 180
181 VGSTFALPRI VTLANSNSQM ALYLCLTGEV VTGADAYMLG LASHYVSSEN LDALQKRLGE 240
241 ISPPFNNDPQ SAYFFGMVNE SIDEFVSPLP KDYVFKYSNE KLNVIEACFN LSKNGTIEDI 300
301 MNNLRQYEGS AEGKAFAQEI KTKLLTKSPS SLQIALRLVQ ENSRDHIESA IKRDLYTAAN 360
361 MCMNQDSLVE FSEATKHKLI DKQRVPYPWT KKEQLFVSQL TSITSPKPSL PMSLLRNTSN 420
421 VTWTQYPYHS KYQLPTEQEI AAYIEKRTND DTGAKVTERE VLNHFANVIP SRRGKLGIQS 480
481 LCKIVCERKC EEVNDGLRWK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
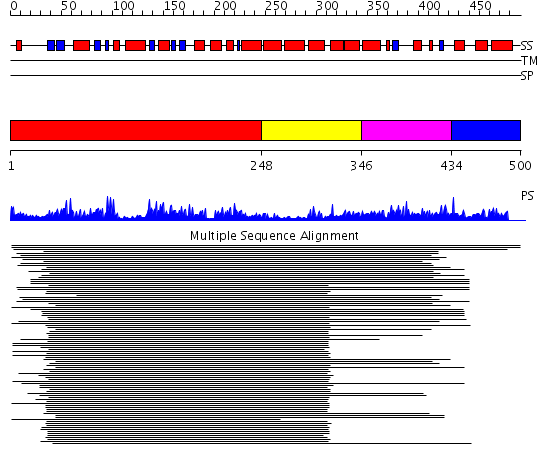
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..247] | 188.201249 | Enoyl-CoA hydratase (crotonase) | |
| 2 | View Details | [248..345] | 188.201249 | Enoyl-CoA hydratase (crotonase) | |
| 3 | View Details | [346..433] | 3.30103 | Short chain L-3-hydroxyacyl CoA dehydrogenase | |
| 4 | View Details | [434..500] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 | |
| 2 | No functions predicted. |
| 3 | No functions predicted. |
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)