













| General Information: |
|
| Name(s) found: |
UTP22 /
YGR090W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1237 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear outer membrane [IMP nucleolus [IDA small nucleolar ribonucleoprotein complex [IPI |
| Biological Process: |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)
[IC]
ribosome biogenesis [IEA] rRNA processing [IEA][IMP] |
| Molecular Function: |
snoRNA binding
[RCA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATSVKRKAS ETSDQNIVKV QKKHSTQDST TDNGSKENDH SSQAINERTV PEQENDESDT 60
61 SPESNEVATN TAATRHNGKV TATESYDIHI ARETAELFKS NIFKLQIDEL LEQVKLKQKH 120
121 VLKVEKFLHK LYDILQEIPD WEEKSLAEVD SFFKNKIVSV PFVDPKPIPQ NTNYKFNYKK 180
181 PDISLIGSFA LKAGIYQPNG SSIDTLLTMP KELFEKKDFL NFRCLHKRSV YLAYLTHHLL 240
241 ILLKKDKLDS FLQLEYSYFD NDPLLPILRI SCSKPTGDSL SDYNFYKTRF SINLLIGFPY 300
301 KVFEPKKLLP NRNCIRIAQE SKEQSLPATP LYNFSVLSSS THENYLKYLY KTKKQTESFV 360
361 EATVLGRLWL QQRGFSSNMS HSGSLGGFGT FEFTILMAAL LNGGGINSNK ILLHGFSSYQ 420
421 LFKGVIKYLA TMDLCHDGHL QFHSNPENSS SSPASKYIDE GFQTPTLFDK STKVNILTKM 480
481 TVSSYQILKE YAGETLRMLN NVVQDQFSNI FLTNISRFDN LKYDLCYDVQ LPLGKYNNLE 540
541 TSLAATFGSM ERVKFITLEN FLAHKITNVA RYALGDRIKY IQIEMVGQKS DFPITKRKVY 600
601 SNTGGNHFNF DFVRVKLIVN PSECDKLVTK GPAHSETMST EAAVFKNFWG IKSSLRRFKD 660
661 GSITHCCVWS TSSSEPIISS IVNFALQKHV SKKAQISNET IKKFHNFLPL PNLPSSAKTS 720
721 VLNLSSFFNL KKSFDDLYKI IFQMKLPLSV KSILPVGSAF RYTSLCQPVP FAYSDPDFFQ 780
781 DVILEFETSP KWPDEITSLE KAKTAFLLKI QEELSANSST YRSFFSRDES IPYNLEIVTL 840
841 NILTPEGYGF KFRVLTERDE ILYLRAIANA RNELKPELEA TFLKFTAKYL ASVRHTRTLE 900
901 NISHSYQFYS PVVRLFKRWL DTHLLLGHIT DELAELIAIK PFVDPAPYFI PGSLENGFLK 960
961 VLKFISQWNW KDDPLILDLV KPEDDIRDTF ETSIGAGSEL DSKTMKKLSE RLTLAQYKGI1020
1021 QMNFTNLRNS DPNGTHLQFF VASKNDPSGI LYSSGIPLPI ATRLTALAKV AVNLLQTHGL1080
1081 NQQTINLLFT PGLKDYDFVV DLRTPIGLKS SCGILSATEF KNITNDQAPS NFPENLNDLS1140
1141 EKMDPTYQLV KYLNLKYKNS LILSSRKYIG VNGGEKGDKN VITGLIKPLF KGAHKFRVNL1200
1201 DCNVKPVDDE NVILNKEAIF HEIAAFGNDM VINFETD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
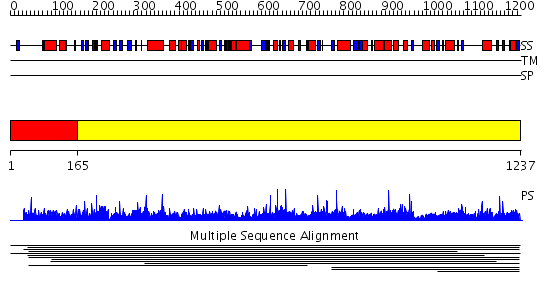
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [165..1237] | 1000.0 | Nrap protein No confident structure predictions are available. | |
| 3 | View Details | [906..1033] | 1.050982 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1034..1237] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)