













| General Information: |
|
| Name(s) found: |
KEM1 /
YGL173C
[SGD]
|
| Description(s) found:
Found 38 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1528 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[TAS cytoplasmic mRNA processing body [IDA nuclear outer membrane [TAS |
| Biological Process: |
filamentous growth
[IMP response to exogenous dsRNA [IMP telomere maintenance [IMP traversing start control point of mitotic cell cycle [IMP karyogamy involved in conjugation with cellular fusion [IGI mRNA catabolic process [TAS |
| Molecular Function: |
5'-3' exoribonuclease activity
[TAS deoxyribonuclease activity [TAS recombinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIPKFFRYI SERWPMILQL IEGTQIPEFD NLYLDMNSIL HNCTHGNDDD VTKRLTEEEV 60
61 FAKICTYIDH LFQTIKPKKI FYMAIDGVAP RAKMNQQRAR RFRTAMDAEK ALKKAIENGD 120
121 EIPKGEPFDS NSITPGTEFM AKLTKNLQYF IHDKISNDSK WREVQIIFSG HEVPGEGEHK 180
181 IMNFIRHLKS QKDFNQNTRH CIYGLDADLI MLGLSTHGPH FALLREEVTF GRRNSEKKSL 240
241 EHQNFYLLHL SLLREYMELE FKEIADEMQF EYNFERILDD FILVMFVIGN DFLPNLPDLH 300
301 LNKGAFPVLL QTFKEALLHT DGYINEHGKI NLKRLGVWLN YLSQFELLNF EKDDIDVEWF 360
361 NKQLENISLE GERKRQRVGK KLLVKQQKKL IGSIKPWLME QLQEKLSPDL PDEEIPTLEL 420
421 PKDLDMKDHL EFLKEFAFDL GLFITHSKSK GSYSLKMDLD SINPDETEEE FQNRVNSIRK 480
481 TIKKYQNAII VEDKEELETE KTIYNERFER WKHEYYHDKL KFTTDSEEKV RDLAKDYVEG 540
541 LQWVLYYYYR GCPSWSWYYP HHYAPRISDL AKGLDQDIEF DLSKPFTPFQ QLMAVLPERS 600
601 KNLIPPAFRP LMYDEQSPIH DFYPAEVQLD KNGKTADWEA VVLISFVDEK RLIEAMQPYL 660
661 RKLSPEEKTR NQFGKDLIYS FNPQVDNLYK SPLGGIFSDI EHNHCVEKEY ITIPLDSSEI 720
721 RYGLLPNAKL GAEMLAGFPT LLSLPFTSSL EYNETMVFQQ PSKQQSMVLQ ITDIYKTNNV 780
781 TLEDFSKRHL NKVIYTRWPY LRESKLVSLT DGKTIYEYQE SNDKKKFGFI TKPAETQDKK 840
841 LFNSLKNSML RMYAKQKAVK IGPMEAIATV FPVTGLVRDS DGGYIKTFSP TPDYYPLQLV 900
901 VESVVNEDER YKERGPIPIE EEFPLNSKVI FLGDYAYGGE TTIDGYSSDR RLKITVEKKF 960
961 LDSEPTIGKE RLQMDHQAVK YYPSYIVSKN MHLHPLFLSK ITSKFMITDA TGKHINVGIP1020
1021 VKFEARHQKV LGYARRNPRG WEYSNLTLNL LKEYRQTFPD FFFRLSKVGN DIPVLEDLFP1080
1081 DTSTKDAMNL LDGIKQWLKY VSSKFIAVSL ESDSLTKTSI AAVEDHIMKY AANIEGHERK1140
1141 QLAKVPREAV LNPRSSFALL RSQKFDLGDR VVYIQDSGKV PIFSKGTVVG YTTLSSSLSI1200
1201 QVLFDHEIVA GNNFGGRLRT NRGLGLDASF LLNITNRQFI YHSKASKKAL EKKKQSNNRN1260
1261 NNTKTAHKTP SKQQSEEKLR KERAHDLLNF IKKDTNEKNS ESVDNKSMGS QKDSKPAKKV1320
1321 LLKRPAQKSS ENVQVDLANF EKAPLDNPTV AGSIFNAVAN QYSDGIGSNL NIPTPPHPMN1380
1381 VVGGPIPGAN DVADVGLPYN IPPGFMTHPN GLHPLHPHQM PYPNMNGMSI PPPAPHGFGQ1440
1441 PISFPPPPPM TNVSDQGSRI VVNEKESQDL KKFINGKQHS NGSTIGGETK NSRKGEIKPS1500
1501 SGTNSTECQS PKSQSNAADR DNKKDEST |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
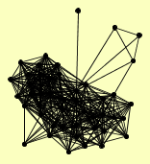
|
ASC1 BRR2 CBF5 DHH1 FUN12 GBP2 IMD3 IMD4 KEM1 LSM1 LSM2 LSM3 LSM4 LSM5 LSM6 LSM7 LSP1 MRP8 NPL3 NRD1 PAT1 PIL1 PRP3 PRP31 PRP4 PRP43 PRP8 PUF6 RPL15A RPL20B RPL6A RPS11A, RPS11B RPS13 RPS4B, RPS4A RPS8B, RPS8A SBP1 SMD3 SNU114 SRO9 STM1 STO1 TIF4631 TIF4632 UTP22 YGR054W YKL023W |
| View Details | Krogan NJ, et al. (2006) |
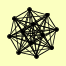
|
KEM1 LSM1 LSM2 LSM3 LSM4 LSM5 LSM6 LSM7 LSM8 PAT1 PRP24 YGR054W |
| View Details | Qiu et al. (2008) |
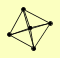
|
DCP1 DCP2 KEM1 LSM1 PAT1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #01 Alpha factor Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #28 Asynchronous Prep5-TiO2 Phosphopeptide Enrichment | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
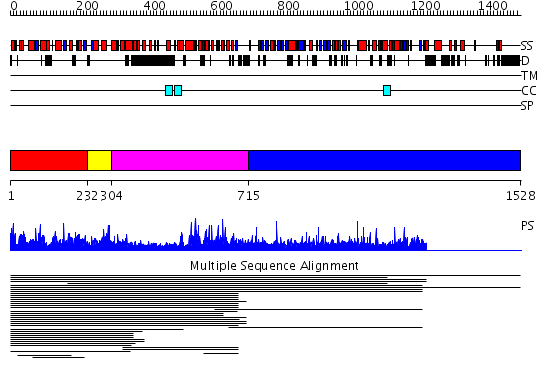
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..231] | 171.387216 | XRN 5'-3' exonuclease N-terminus No confident structure predictions are available. | |
| 2 | View Details | [232..303] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [304..714] | 1.021889 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [715..1528] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [865..938] | 1.034995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [939..1146] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1147..1221] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1222..1528] | 1.23 | THE BLUETONGUE VIRUS (BTV) CORE BINDS DSRNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)