













| General Information: |
|
| Name(s) found: |
PRP43 /
YGL120C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA spliceosomal complex [TAS nuclear outer membrane [IMP |
| Biological Process: |
maturation of SSU-rRNA
[IMP ribosomal large subunit biogenesis [IMP ribosome biogenesis [RCA rRNA processing [IMP processing of 27S pre-rRNA [IMP |
| Molecular Function: |
ATP-dependent RNA helicase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSKRRFSSE HPDPVETSIP EQAAEIAEEL SKQHPLPSEE PLVHHDAGEF KGLQRHHTSA 60
61 EEAQKLEDGK INPFTGREFT PKYVDILKIR RELPVHAQRD EFLKLYQNNQ IMVFVGETGS 120
121 GKTTQIPQFV LFDEMPHLEN TQVACTQPRR VAAMSVAQRV AEEMDVKLGE EVGYSIRFEN 180
181 KTSNKTILKY MTDGMLLREA MEDHDLSRYS CIILDEAHER TLATDILMGL LKQVVKRRPD 240
241 LKIIIMSATL DAEKFQRYFN DAPLLAVPGR TYPVELYYTP EFQRDYLDSA IRTVLQIHAT 300
301 EEAGDILLFL TGEDEIEDAV RKISLEGDQL VREEGCGPLS VYPLYGSLPP HQQQRIFEPA 360
361 PESHNGRPGR KVVISTNIAE TSLTIDGIVY VVDPGFSKQK VYNPRIRVES LLVSPISKAS 420
421 AQQRAGRAGR TRPGKCFRLY TEEAFQKELI EQSYPEILRS NLSSTVLELK KLGIDDLVHF 480
481 DFMDPPAPET MMRALEELNY LACLDDEGNL TPLGRLASQF PLDPMLAVML IGSFEFQCSQ 540
541 EILTIVAMLS VPNVFIRPTK DKKRADDAKN IFAHPDGDHI TLLNVYHAFK SDEAYEYGIH 600
601 KWCRDHYLNY RSLSAADNIR SQLERLMNRY NLELNTTDYE SPKYFDNIRK ALASGFFMQV 660
661 AKKRSGAKGY ITVKDNQDVL IHPSTVLGHD AEWVIYNEFV LTSKNYIRTV TSVRPEWLIE 720
721 IAPAYYDLSN FQKGDVKLSL ERIKEKVDRL NELKQGKNKK KSKHSKK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
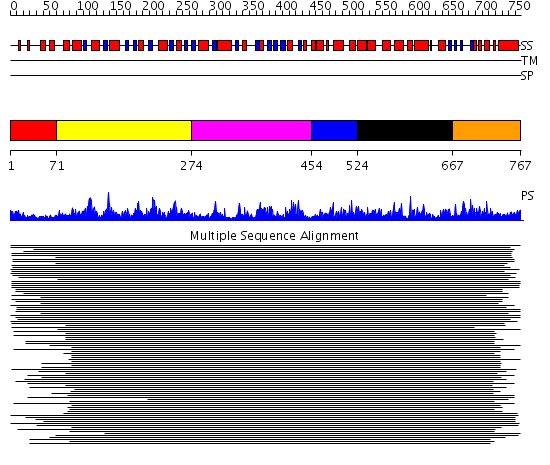
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [71..273] | 61.69897 | Putative DEAD box RNA helicase | |
| 3 | View Details | [274..453] | 61.69897 | Putative DEAD box RNA helicase | |
| 4 | View Details | [454..523] | 7.23 | delta subunit; delta subunit of DNA polymerase III, N-domain | |
| 5 | View Details | [524..666] | 7.23 | delta subunit; delta subunit of DNA polymerase III, N-domain | |
| 6 | View Details | [667..767] | 3.157986 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)