













| General Information: |
|
| Name(s) found: |
PRP42 /
YDR235W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 544 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U1 snRNP
[IDA |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IMP |
| Molecular Function: |
RNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKYTALIHD ENFSTLTLNV SRYPKSLAYW EKLLNYIVKA SAPICKSTEP QLLKLIRCTY 60
61 SSMLNEFPYL ENYYIDFALL EYKLGNVSMS HKIFQRGLQA FNQRSLLLWT SYLKFCNNVI 120
121 SHQKQLFKKY ETAEEYVGLH FFSGEFWDLY LEQISSRCTS SKKYWNVLRK ILEIPLHSFS 180
181 KFYALWLQRI DDIMDLKQLS QLTSKDELLK KLKIDINYSG RKGPYLQDAK KKLKKITKEM 240
241 YMVVQYQVLE IYSIFESKIY INYYTSPETL VSSDEIETWI KYLDYTITLQ TDSLTHLNFQ 300
301 RALLPLAHYD LVWIKYSKWL INSKNDLLGA KNVLLMGLKF SLKKTEIIKL LYSVICKLNE 360
361 YVLLRNLLEK IESSYSDNVE NVDDFEIFWD YLQFKTFCQN SLYSSRYSDS QSNGLLNKEL 420
421 FDKVWKRLSC KEKKSGQEIL LNNLVQFYSK DTVEFVEKNI FQKIIEFGWE YYLQNGMFWN 480
481 CYCRLIYFDT SRSYLDKRQY IVRKIWPQID KKFAQSVLPS LTEFCESYFP EEMDTLEEMF 540
541 TEEP |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
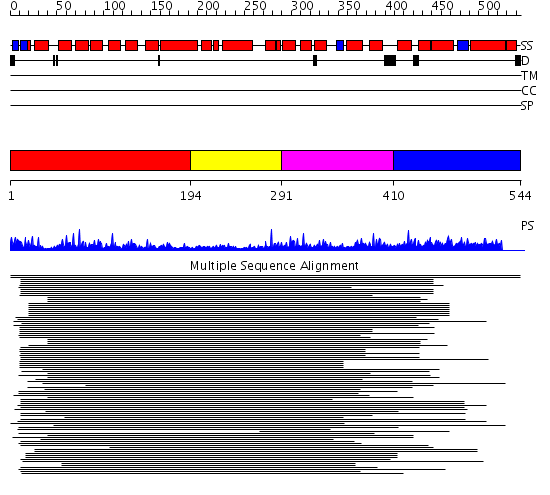
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..193] | 16.045757 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 2 | View Details | [194..290] | 16.045757 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 3 | View Details | [291..409] | 16.045757 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 4 | View Details | [410..544] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)