













| General Information: |
|
| Name(s) found: |
YEF3 /
YLR249W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1044 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ribosome
[IDA |
| Biological Process: |
ribosome biogenesis
[RCA translational elongation [TAS |
| Molecular Function: |
translation elongation factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDSQQSIKV LEELFQKLSV ATADNRHEIA SEVASFLNGN IIEHDVPEHF FGELAKGIKD 60
61 KKTAANAMQA VAHIANQSNL SPSVEPYIVQ LVPAICTNAG NKDKEIQSVA SETLISIVNA 120
121 VNPVAIKALL PHLTNAIVET NKWQEKIAIL AAISAMVDAA KDQVALRMPE LIPVLSETMW 180
181 DTKKEVKAAA TAAMTKATET VDNKDIERFI PSLIQCIADP TEVPETVHLL GATTFVAEVT 240
241 PATLSIMVPL LSRGLNERET GIKRKSAVII DNMCKLVEDP QVIAPFLGKL LPGLKSNFAT 300
301 IADPEAREVT LRALKTLRRV GNVGEDDAIP EVSHAGDVST TLQVVNELLK DETVAPRFKI 360
361 VVEYIAAIGA DLIDERIIDQ QAWFTHITPY MTIFLHEKKA KDILDEFRKR AVDNIPVGPN 420
421 FDDEEDEGED LCNCEFSLAY GAKILLNKTQ LRLKRARRYG ICGPNGCGKS TLMRAIANGQ 480
481 VDGFPTQEEC RTVYVEHDID GTHSDTSVLD FVFESGVGTK EAIKDKLIEF GFTDEMIAMP 540
541 ISALSGGWKM KLALARAVLR NADILLLDEP TNHLDTVNVA WLVNYLNTCG ITSITISHDS 600
601 VFLDNVCEYI INYEGLKLRK YKGNFTEFVK KCPAAKAYEE LSNTDLEFKF PEPGYLEGVK 660
661 TKQKAIVKVT NMEFQYPGTS KPQITDINFQ CSLSSRIAVI GPNGAGKSTL INVLTGELLP 720
721 TSGEVYTHEN CRIAYIKQHA FAHIESHLDK TPSEYIQWRF QTGEDRETMD RANRQINEND 780
781 AEAMNKIFKI EGTPRRIAGI HSRRKFKNTY EYECSFLLGE NIGMKSERWV PMMSVDNAWI 840
841 PRGELVESHS KMVAEVDMKE ALASGQFRPL TRKEIEEHCS MLGLDPEIVS HSRIRGLSGG 900
901 QKVKLVLAAG TWQRPHLIVL DEPTNYLDRD SLGALSKALK EFEGGVIIIT HSAEFTKNLT 960
961 EEVWAVKDGR MTPSGHNWVS GQGAGPRIEK KEDEEDKFDA MGNKIAGGKK KKKLSSAELR1020
1021 KKKKERMKKK KELGDAYVSS DEEF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ABP1 ADH1 ADH3 CAM1 EFB1 EFT2, EFT1 FBA1 GUS1 HSC82 HSP104 HSP82 HXK1 HYP2 PDC1 PIL1 RPG1 RPL40B, RPL40A SSZ1 TDH2 TEF4 TIF1, TIF2 YEF3 ZUO1 |
| View Details | Krogan NJ, et al. (2006) |
|
HFM1 LYS4 NEW1 RBG1 SGD1 SOK1 SSZ1 TCB1 TGL5 TMA46 YEF3 YLR072W ZUO1 |
| View Details | Gavin AC, et al. (2002) |
|
ACT1 ADH1 AOS1 ARX1 BGL2 BRR1 CBC2 CDC14 CDC33 CLF1 CSN12 CUS1 CWC2 DIB1 ECM2 GCN1 GIN4 HMO1 HSC82 HSH155 HSP42 ISW1 KAP114 KAP123 KCC4 LEA1 LSM4 LUC7 MLC1 MLC2 MOT1 MSL1 MUD1 MYO1 MYO2 MYO4 NAM8 NET1 NIP1 NPL3 PBP2 PDC1 PFK2 PGI1 PRP11 PRP19 PRP21 PRP3 PRP31 PRP39 PRP4 PRP40 PRP42 PRP43 PRP46 PRP6 PRP9 RAD1 RAD10 RPN9 RPT5 RSE1 SHE2 SHE3 SIR2 SIR4 SMB1 SMD1 SMD2 SMD3 SME1 SMX2 SMX3 SNP1 SNT309 SNU114 SNU56 SNU66 SNU71 SPP382 SPT15 SRB2 STO1 TAF1 TAF5 TAF6 TDH3 TIF4631 TIF4632 UBA2 VPS1 YEF3 YRA1 |
| View Details | Qiu et al. (2008) |

|
TEF1, TEF2 YEF3 |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
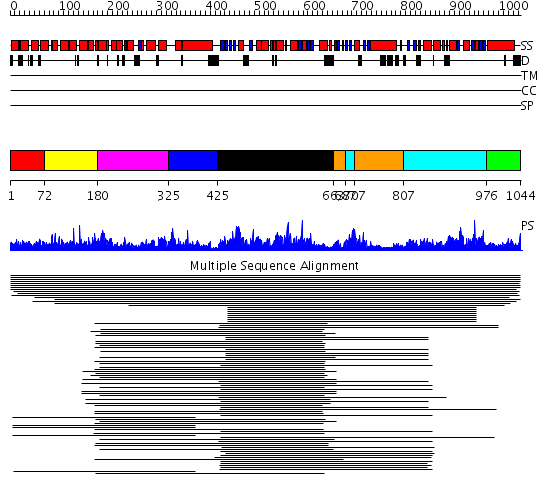
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | 13.93 | Lipovitellin-phosvitin complex, superhelical domain; Lipovitellin-phosvitin complex; beta-sheet shell regions | |
| 2 | View Details | [72..179] | 13.93 | Lipovitellin-phosvitin complex, superhelical domain; Lipovitellin-phosvitin complex; beta-sheet shell regions | |
| 3 | View Details | [180..324] | 13.93 | Lipovitellin-phosvitin complex, superhelical domain; Lipovitellin-phosvitin complex; beta-sheet shell regions | |
| 4 | View Details | [325..424] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [425..662] | 105.0 | Maltose transport protein MalK, C-terminal domain; Maltose transport protein MalK, N-terminal domain | |
| 6 | View Details | [663..686] [707..806] |
4.69897 | Smc head domain | |
| 7 | View Details | [687..706] [807..975] |
4.69897 | Smc head domain | |
| 8 | View Details | [976..1044] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)