













| General Information: |
|
| Name(s) found: |
PRP9 /
YDL030W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 530 amino acids |
Gene Ontology: |
|
| Cellular Component: |
U2 snRNP
[IDA |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IDA |
| Molecular Function: |
RNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNLLETRRSL LEEMEIIENA IAERIQRNPE LYYHYIQESS KVFPDTKLPR SSLIAENKIY 60
61 KFKKVKRKRK QIILQQHEIN IFLRDYQEKQ QTFNKINRPE ETQEDDKDLP NFERKLQQLE 120
121 KELKNEDENF ELDINSKKDK YALFSSSSDP SRRTNILSDR ARDLDLNEIF TRDEQYGEYM 180
181 ELEQFHSLWL NVIKRGDCSL LQFLDILELF LDDEKYLLTP PMDRKNDRYM AFLLKLSKYV 240
241 ETFFFKSYAL LDAAAVENLI KSDFEHSYCR GSLRSEAKGI YCPFCSRWFK TSSVFESHLV 300
301 GKIHKKNESK RRNFVYSEYK LHRYLKYLND EFSRTRSFVE RKLAFTANER MAEMDILTQK 360
361 YEAPAYDSTE KEGAEQVDGE QRDGQLQEEH LSGKSFDMPL GPDGLPMPYW LYKLHGLDRE 420
421 YRCEICSNKV YNGRRTFERH FNEERHIYHL RCLGIEPSSV FKGITKIKEA QELWKNMQGQ 480
481 SQLTSIAAVP PKPNPSQLKV PTELELEEED EEGNVMSKKV YDELKKQGLV |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #20 Asynchronous Prep1-TiO2 Phosphopeptide enrichment, PartB | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
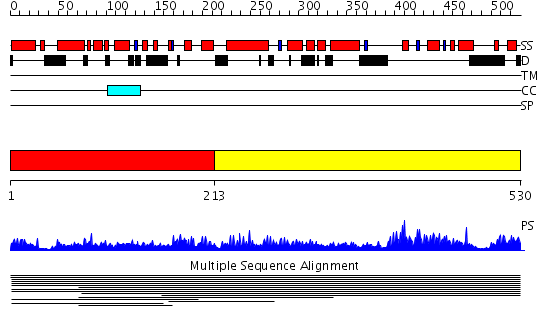
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..212] | 1.012986 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [213..530] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)