













| General Information: |
|
| Name(s) found: |
PRP19 /
YLL036C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 503 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA mitochondrion [IDA spliceosomal complex [IDA |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IDA ubiquitin-dependent protein catabolic process [IPI |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA][IEA][IMP]
first spliceosomal transesterification activity [IC] RNA binding [RCA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLCAISGKVP RRPVLSPKSR TIFEKSLLEQ YVKDTGNDPI TNEPLSIEEI VEIVPSAQQA 60
61 SLTESTNSAT LKANYSIPNL LTSLQNEWDA IMLENFKLRS TLDSLTKKLS TVMYERDAAK 120
121 LVAAQLLMEK NEDSKDLPKS SQQAVAITRE EFLQGLLQSS RDFVARGKLK APKWPILKNL 180
181 ELLQAQNYSR NIKTFPYKEL NKSMYYDKWV CMCRCEDGAL HFTQLKDSKT ITTITTPNPR 240
241 TGGEHPAIIS RGPCNRLLLL YPGNQITILD SKTNKVLREI EVDSANEIIY MYGHNEVNTE 300
301 YFIWADNRGT IGFQSYEDDS QYIVHSAKSD VEYSSGVLHK DSLLLALYSP DGILDVYNLS 360
361 SPDQASSRFP VDEEAKIKEV KFADNGYWMV VECDQTVVCF DLRKDVGTLA YPTYTIPEFK 420
421 TGTVTYDIDD SGKNMIAYSN ESNSLTIYKF DKKTKNWTKD EESALCLQSD TADFTDMDVV 480
481 CGDGGIAAIL KTNDSFNIVA LTP |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
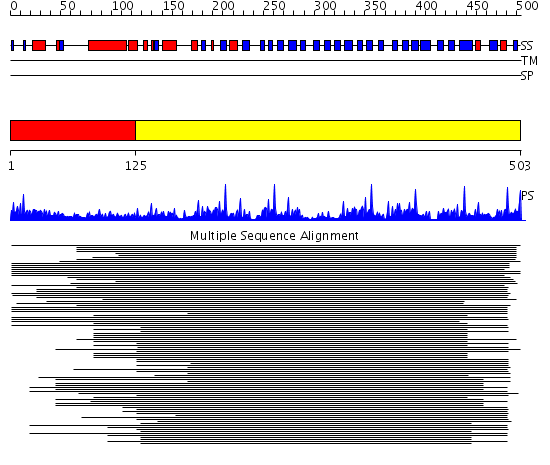
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..124] | 2.69897 | STAT-1, coiled coil domain; STAT-1, DNA-binding domain; STAT-1 | |
| 2 | View Details | [125..503] | 77.39794 | beta1-subunit of the signal-transducing G protein heterotrimer |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)