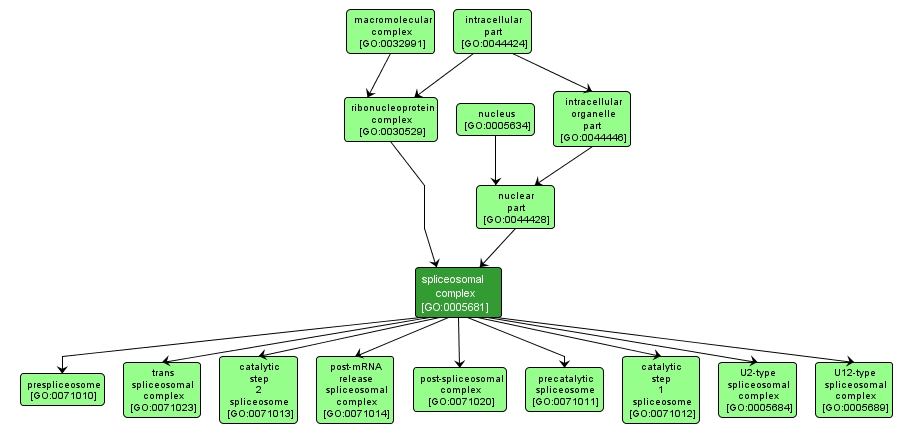
GO TERM SUMMARY
|
| Name: |
spliceosomal complex |
| Acc: |
GO:0005681 |
| Aspect: |
Cellular Component |
| Desc: |
Any of a series of ribonucleoprotein complexes that contain RNA and small nuclear ribonucleoproteins (snRNPs), and are formed sequentially during the splicing of a messenger RNA primary transcript to excise an intron. |
| Synonyms:
|
|

|
INTERACTIVE GO GRAPH
|