













| General Information: |
|
| Name(s) found: |
SNP1 /
YIL061C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 300 amino acids |
Gene Ontology: |
|
| Cellular Component: |
commitment complex
[IPI U1 snRNP [IDA |
| Biological Process: |
nuclear mRNA splicing, via spliceosome
[IPI |
| Molecular Function: |
U1 snRNA binding
[IPI mRNA binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNYNLSKYPD DVSRLFKPRP PLSYKRPTDY PYAKRQTNPN ITGVANLLST SLKHYMEEFP 60
61 EGSPNNHLQR YEDIKLSKIK NAQLLDRRLQ NWNPNVDPHI KDTDPYRTIF IGRLPYDLDE 120
121 IELQKYFVKF GEIEKIRIVK DKITQKSKGY AFIVFKDPIS SKMAFKEIGV HRGIQIKDRI 180
181 CIVDIERGRT VKYFKPRRLG GGLGGRGYSN RDSRLPGRFA SASTSNPAER NYAPRLPRRE 240
241 TSSSAYSADR YGSSTLDARY RGNRPLLSAA TPTAAVTSVY KSRNSRTRES QPAPKEAPDY 300
301 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #27 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps3-5 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
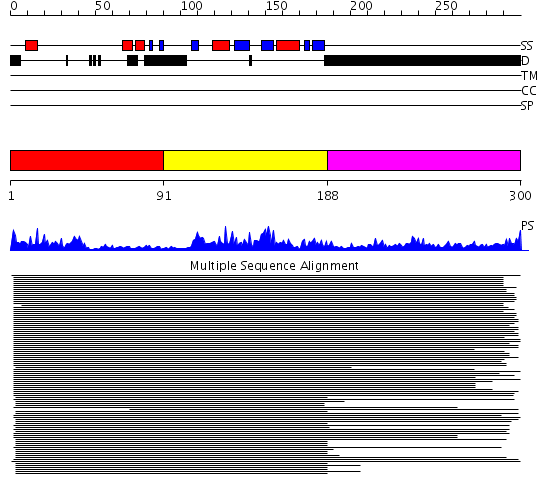
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 99.9691 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 2 | View Details | [91..187] | 99.9691 | Nuclear ribonucleoprotein A1 (RNP A1, UP1) | |
| 3 | View Details | [188..300] | 80.30103 | Hu antigen D (Hud) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)