













| General Information: |
|
| Name(s) found: |
HEK2 /
YBL032W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 381 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA nuclear chromosome, telomeric region [IDA |
| Biological Process: |
intracellular mRNA localization
[IMP telomere maintenance via telomerase [IMP |
| Molecular Function: |
mRNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQFFEAATP VAIPTNNTNG GSSDAGSAAT GGAPVVGTTA QPTINHRLLL SLKEAAKIIG 60
61 TKGSTISRIR AANAVKIGIS EKVPGCSDRI LSCAGNVINV ANAIGDIVDV LNKRNPENED 120
121 AAEGEAEEHY YFHFLNHILP APSKDEIRDL QQLEDIGYVR LIVANSHISS IIGKAGATIK 180
181 SLINKHGVKI VASKDFLPAS DERIIEIQGF PGSITNVLIE ISEIILSDVD VRFSTERSYF 240
241 PHLKKSSGEP TSPSTSSNTR IELKIPELYV GAIIGRGMNR IKNLKTFTKT NIVVERKDDD 300
301 DKDENFRKFI ITSKFPKNVK LAESMLLKNL NTEIEKRENY KRKLEAAEGD ATVVTERSDS 360
361 ASFLEEKEEP QENHDNKEEQ S |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ARC1 ATG14 CAT8 CCC2 HEK2 IRC20 MCA1 MES1 MGA2 NCS2 NHA1 PET127 PHO89 PHO91 YKL050C YLR363W-A |
| View Details | Gavin AC, et al. (2002) |

|
ASC1 CBF5 CDC33 DBP2 HEK2 HHF1, HHF2 LHP1 MIS1 MRP7 NAB6 POL2 PUS7 RBG1 ROM2 SBP1 SNP1 SRO9 STM1 STO1 SUI3 TIF1, TIF2 TIF4631 TIF4632 UTP20 YMR310C |
| View Details | Qiu et al. (2008) |
|
HEK2 RAP1 RIF1 SUB2 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
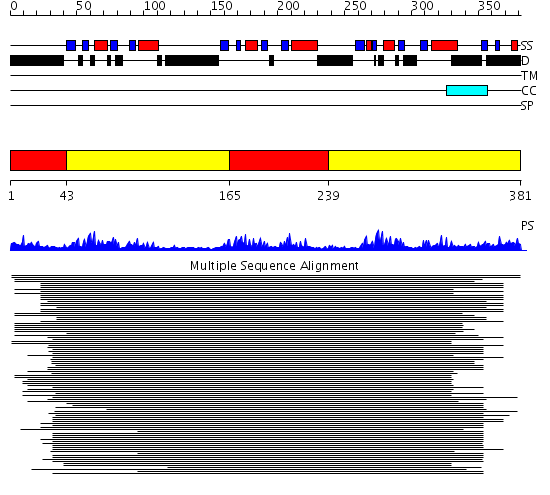
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..42] [165..238] |
36.30103 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 | |
| 2 | View Details | [43..164] [239..381] |
36.30103 | Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 3; S1 RNA-binding domain of polyribonucleotide phosphorylase, PNPase; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 1 and 4; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domain 6; Polynucleotide phosphorylase/guanosine pentaphosphate synthase (PNPase/GPSI), domains 2 and 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)