













| General Information: |
|
| Name(s) found: |
POL2 /
YNL262W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2222 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[IDA epsilon DNA polymerase complex [IDA |
| Biological Process: |
lagging strand elongation
[TAS leading strand elongation [TAS nucleotide-excision repair [TAS mismatch repair [TAS DNA synthesis involved in DNA repair [IMP chromatin silencing at telomere [IMP |
| Molecular Function: |
nucleotide binding
[IEA]
nucleic acid binding [IEA] DNA binding [IEA] zinc ion binding [IEA] DNA-directed DNA polymerase activity [IEA][IMP] nucleotidyltransferase activity [IEA] metal ion binding [IEA] double-stranded DNA binding [IDA] transferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMFGKKKNNG GSSTARYSAG NKYNTLSNNY ALSAQQLLNA SKIDDIDSMM GFERYVPPQY 60
61 NGRFDAKDID QIPGRVGWLT NMHATLVSQE TLSSGSNGGG NSNDGERVTT NQGISGVDFY 120
121 FLDEEGGSFK STVVYDPYFF IACNDESRVN DVEELVKKYL ESCLKSLQII RKEDLTMDNH 180
181 LLGLQKTLIK LSFVNSNQLF EARKLLRPIL QDNANNNVQR NIYNVAANGS EKVDAKHLIE 240
241 DIREYDVPYH VRVSIDKDIR VGKWYKVTQQ GFIEDTRKIA FADPVVMAFD IETTKPPLKF 300
301 PDSAVDQIMM ISYMIDGEGF LITNREIISE DIEDFEYTPK PEYPGFFTIF NENDEVALLQ 360
361 RFFEHIRDVR PTVISTFNGD FFDWPFIHNR SKIHGLDMFD EIGFAPDAEG EYKSSYCSHM 420
421 DCFRWVKRDS YLPQGSQGLK AVTQSKLGYN PIELDPELMT PYAFEKPQHL SEYSVSDAVA 480
481 TYYLYMKYVH PFIFSLCTII PLNPDETLRK GTGTLCEMLL MVQAYQHNIL LPNKHTDPIE 540
541 RFYDGHLLES ETYVGGHVES LEAGVFRSDL KNEFKIDPSA IDELLQELPE ALKFSVEVEN 600
601 KSSVDKVTNF EEIKNQITQK LLELKENNIR NELPLIYHVD VASMYPNIMT TNRLQPDSIK 660
661 AERDCASCDF NRPGKTCARK LKWAWRGEFF PSKMDEYNMI KRALQNETFP NKNKFSKKKV 720
721 LTFDELSYAD QVIHIKKRLT EYSRKVYHRV KVSEIVEREA IVCQRENPFY VDTVKSFRDR 780
781 RYEFKGLAKT WKGNLSKIDP SDKHARDEAK KMIVLYDSLQ LAHKVILNSF YGYVMRKGSR 840
841 WYSMEMAGIT CLTGATIIQM ARALVERVGR PLELDTDGIW CILPKSFPET YFFTLENGKK 900
901 LYLSYPCSML NYRVHQKFTN HQYQELKDPL NYIYETHSEN TIFFEVDGPY KAMILPSSKE 960
961 EGKGIKKRYA VFNEDGSLAE LKGFELKRRG ELQLIKNFQS DIFKVFLEGD TLEGCYSAVA1020
1021 SVCNRWLDVL DSHGLMLEDE DLVSLICENR SMSKTLKEYE GQKSTSITTA RRLGDFLGED1080
1081 MVKDKGLQCK YIISSKPFNA PVTERAIPVA IFSADIPIKR SFLRRWTLDP SLEDLDIRTI1140
1141 IDWGYYRERL GSAIQKIITI PAALQGVSNP VPRVEHPDWL KRKIATKEDK FKQTSLTKFF1200
1201 SKTKNVPTMG KIKDIEDLFE PTVEEDNAKI KIARTTKKKA VSKRKRNQLT NEEDPLVLPS1260
1261 EIPSMDEDYV GWLNYQKIKW KIQARDRKRR DQLFGNTNSS RERSALGSMI RKQAESYANS1320
1321 TWEVLQYKDS GEPGVLEVFV TINGKVQNIT FHIPKTIYMK FKSQTMPLQK IKNCLIEKSS1380
1381 ASLPNNPKTS NPAGGQLFKI TLPESVFLEE KENCTSIFND ENVLGVFEGT ITPHQRAIMD1440
1441 LGASVTFRSK AMGALGKGIQ QGFEMKDLSM AENERYLSGF SMDIGYLLHF PTSIGYEFFS1500
1501 LFKSWGDTIT ILVLKPSNQA QEINASSLGQ IYKQMFEKKK GKIETYSYLV DIKEDINFEF1560
1561 VYFTDISKLY RRLSQETTKL KEERGLQFLL LLQSPFITKL LGTIRLLNQM PIVKLSLNEV1620
1621 LLPQLNWQPT LLKKLVNHVL SSGSWISHLI KLSQYSNIPI CNLRLDSMDY IIDVLYARKL1680
1681 KKENIVLWWN EKAPLPDHGG IQNDFDLNTS WIMNDSEFPK INNSGVYDNV VLDVGVDNLT1740
1741 VNTILTSALI NDAEGSDLVN NNMGIDDKDA VINSPSEFVH DAFSNDALNV LRGMLKEWWD1800
1801 EALKENSTAD LLVNSLASWV QNPNAKLFDG LLRYHVHNLT KKALLQLVNE FSALGSTIVY1860
1861 ADRNQILIKT NKYSPENCYA YSQYMMKAVR TNPMFSYLDL NIKRYWDLLI WMDKFNFSGL1920
1921 ACIEIEEKEN QDYTAVSQWQ LKKFLSPIYQ PEFEDWMMII LDSMLKTKQS YLKLNSGTQR1980
1981 PTQIVNVKKQ DKEDSVENSL NGFSHLFSKP LMKRVKKLFK NQQEFILDPQ YEADYVIPVL2040
2041 PGSHLNVKNP LLELVKSLCH VMLLSKSTIL EIRTLRKELL KIFELREFAK VAEFKDPSLS2100
2101 LVVPDFLCEY CFFISDIDFC KAAPESIFSC VRCHKAFNQV LLQEHLIQKL RSDIESYLIQ2160
2161 DLRCSRCHKV KRDYMSAHCP CAGAWEGTLP RESIVQKLNV FKQVAKYYGF DILLSCIADL2220
2221 TI |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
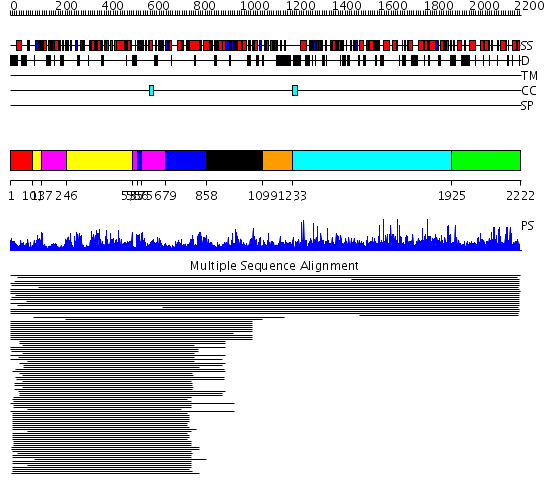
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [101..136] [246..534] |
274.0 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 3 | View Details | [137..245] [535..555] [575..678] |
274.0 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 4 | View Details | [556..574] [679..857] |
274.0 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 5 | View Details | [858..1098] | 13.0 | No description for 1b1fR_ was found. | |
| 6 | View Details | [1099..1232] | 2.037994 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1233..1924] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1925..2222] | 3.018912 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)