













| General Information: |
|
| Name(s) found: |
MCM6 /
YGL201C
[SGD]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1017 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA pre-replicative complex [IDA cytoplasm [IDA MCM complex [IDA |
| Biological Process: |
DNA replication initiation
[IPI DNA unwinding involved in replication [TAS |
| Molecular Function: |
chromatin binding
[TAS ATP-dependent DNA helicase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSPFPADTP SSNRPSNSSP PPSSIGAGFG SSSGLDSQIG SRLHFPSSSQ PHVSNSQTGP 60
61 FVNDSTQFSS QRLQTDGSAT NDMEGNEPAR SFKSRALNHV KKVDDVTGEK VREAFEQFLE 120
121 DFSVQSTDTG EVEKVYRAQI EFMKIYDLNT IYIDYQHLSM RENGALAMAI SEQYYRFLPF 180
181 LQKGLRRVVR KYAPELLNTS DSLKRSEGDE GQADEDEQQD DDMNGSSLPR DSGSSAAPGN 240
241 GTSAMATRSI TTSTSPEQTE RVFQISFFNL PTVHRIRDIR SEKIGSLLSI SGTVTRTSEV 300
301 RPELYKASFT CDMCRAIVDN VEQSFKYTEP TFCPNPSCEN RAFWTLNVTR SRFLDWQKVR 360
361 IQENANEIPT GSMPRTLDVI LRGDSVERAK PGDRCKFTGV EIVVPDVTQL GLPGVKPSST 420
421 LDTRGISKTT EGLNSGVTGL RSLGVRDLTY KISFLACHVI SIGSNIGASS PDANSNNRET 480
481 ELQMAANLQA NNVYQDNERD QEVFLNSLSS DEINELKEMV KDEHIYDKLV RSIAPAVFGH 540
541 EAVKKGILLQ MLGGVHKSTV EGIKLRGDIN ICVVGDPSTS KSQFLKYVVG FAPRSVYTSG 600
601 KASSAAGLTA AVVRDEEGGD YTIEAGALML ADNGICCIDE FDKMDISDQV AIHEAMEQQT 660
661 ISIAKAGIHA TLNARTSILA AANPVGGRYN RKLSLRGNLN MTAPIMSRFD LFFVILDDCN 720
721 EKIDTELASH IVDLHMKRDE AIEPPFSAEQ LRRYIKYART FKPILTKEAR SYLVEKYKEL 780
781 RKDDAQGFSR SSYRITVRQL ESMIRLSEAI ARANCVDEIT PSFIAEAYDL LRQSIIRVDV 840
841 DDVEMDEEFD NIESQSHAAS GNNDDNDDGT GSGVITSEPP ADIEEGQSEA TARPGTSEKK 900
901 KTTVTYDKYV SMMNMIVRKI AEVDREGAEE LTAVDIVDWY LLQKENDLGS LAEYWEERRL 960
961 AFKVIKRLVK DRILMEIHGT RHNLRDLENE ENENNKTVYV IHPNCEVLDQ LEPQDSS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
CDC45 CDC6 DPB11 DPB2 DPB3 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 POL2 POL3 POL31 POL32 SLD3 |
| View Details | Krogan NJ, et al. (2006) |

|
ACK1 CIN8 DCG1 GNA1 MCM6 MLP1 PMD1 PMT4 PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RFT1 SCL1 SGA1 SNQ2 TOS8 UMP1 YCR076C YDR179W-A YJL132W YKR070W YLR211C YLR290C |
| View Details | Gavin AC, et al. (2002) |

|
CDC45 ERG26 GLT1 HAT1 HSP42 LYS12 MCM2 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 RPD3 RPN9 SIN3 TFP1 |
| View Details | Qiu et al. (2008) |

|
ABF1 CDC45 CDC6 DPB11 DPB2 DPB3 DPB4 HCS1 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 PCC1 POB3 POL1 POL12 POL2 POL3 POL30 POL31 POL32 PRI1 RFA1 RFA2 RFA3 RFC1 RFC2 RFC3 RFC4 RRM3 SLD3 TBF1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
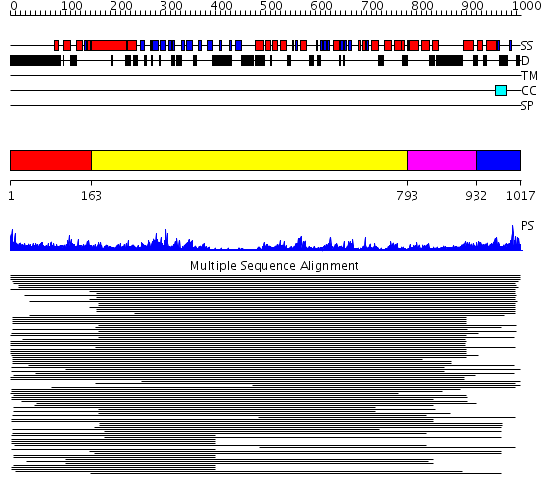
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..162] | 1.107993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [163..792] | 39.09691 | Crystal Structure Analysis of ClpB | |
| 3 | View Details | [793..931] | 19.221849 | Crystal structure of helicobacter pylori ClpX | |
| 4 | View Details | [932..1017] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)