













| General Information: |
|
| Name(s) found: |
POL30 /
YBR088C
[SGD]
|
| Description(s) found:
Found 43 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 258 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[TAS PCNA complex [TAS nucleus [IDA |
| Biological Process: |
mutagenesis
[IGI lagging strand elongation [IDA base-excision repair [TAS leading strand elongation [IDA nucleotide-excision repair [IMP mismatch repair [IMP chromatin silencing at silent mating-type cassette [IMP chromatin silencing at telomere [IMP postreplication repair [IMP |
| Molecular Function: |
DNA polymerase processivity factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLEAKFEEAS LFKRIIDGFK DCVQLVNFQC KEDGIIAQAV DDSRVLLVSL EIGVEAFQEY 60
61 RCDHPVTLGM DLTSLSKILR CGNNTDTLTL IADNTPDSII LLFEDTKKDR IAEYSLKLMD 120
121 IDADFLKIEE LQYDSTLSLP SSEFSKIVRD LSQLSDSINI MITKETIKFV ADGDIGSGSV 180
181 IIKPFVDMEH PETSIKLEMD QPVDLTFGAK YLLDIIKGSS LSDRVGIRLS SEAPALFQFD 240
241 LKSGFLQFFL APKFNDEE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
POL3 POL30 POL31 POL32 |
| View Details | Ho Y, et al. (2002) |
|
CDC9 CYS4 DBP9 ECM10 FPR4 IMD4 MKK2 POL30 RAD27 RPF2 RPO31 YEN1 YOR378W |
| View Details | Qiu et al. (2008) |
|
CDC45 DPB11 DPB2 DPB3 DPB4 MCM2 MCM4 MCM6 MRC1 POL1 POL12 POL2 POL3 POL30 POL31 POL32 PRI1 PRI2 RAD30 RFC4 SLD2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Sundin BA, et al. (2004) |
| View Data |
|
Huh WK, et al. (2003) |
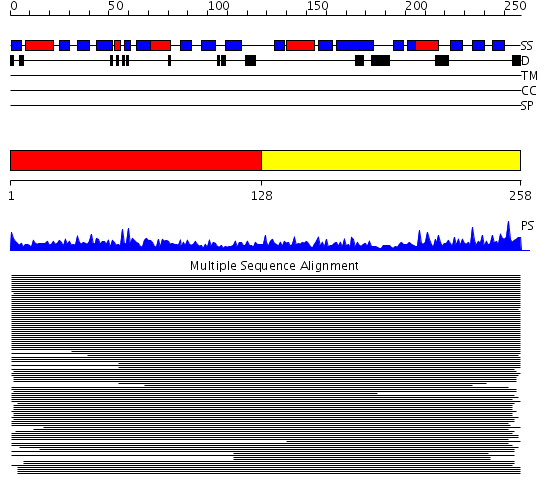
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | 1391.09691 | Prolifirating cell nuclear antigen (PCNA) | |
| 2 | View Details | [128..258] | 1391.09691 | Prolifirating cell nuclear antigen (PCNA) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)