













| General Information: |
|
| Name(s) found: |
CDC9 /
YDL164C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 755 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[NAS nucleus [IDA mitochondrion [IDA |
| Biological Process: |
lagging strand elongation
[IDA base-excision repair [IMP DNA ligation [IDA DNA recombination [IMP nucleotide-excision repair [IMP |
| Molecular Function: |
DNA ligase (ATP) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRRLLTGCLL SSARPLKSRL PLLMSSSLPS SAGKKPKQAT LARFFTSMKN KPTEGTPSPK 60
61 KSSKHMLEDR MDNVSGEEEY ATKKLKQTAV THTVAAPSSM GSNFSSIPSS APSSGVADSP 120
121 QQSQRLVGEV EDALSSNNND HYSSNIPYSE VCEVFNKIEA ISSRLEIIRI CSDFFIKIMK 180
181 QSSKNLIPTT YLFINRLGPD YEAGLELGLG ENLLMKTISE TCGKSMSQIK LKYKDIGDLG 240
241 EIAMGARNVQ PTMFKPKPLT VGEVFKNLRA IAKTQGKDSQ LKKMKLIKRM LTACKGIEAK 300
301 FLIRSLESKL RIGLAEKTVL ISLSKALLLH DENREDSPDK DVPMDVLESA QQKIRDAFCQ 360
361 VPNYEIVINS CLEHGIMNLD KYCTLRPGIP LKPMLAKPTK AINEVLDRFQ GETFTSEYKY 420
421 DGERAQVHLL NDGTMRIYSR NGENMTERYP EINITDFIQD LDTTKNLILD CEAVAWDKDQ 480
481 GKILPFQVLS TRKRKDVELN DVKVKVCLFA FDILCYNDER LINKSLKERR EYLTKVTKVV 540
541 PGEFQYATQI TTNNLDELQK FLDESVNHSC EGLMVKMLEG PESHYEPSKR SRNWLKLKKD 600
601 YLEGVGDSLD LCVLGAYYGR GKRTGTYGGF LLGCYNQDTG EFETCCKIGT GFSDEMLQLL 660
661 HDRLTPTIID GPKATFVFDS SAEPDVWFEP TTLFEVLTAD LSLSPIYKAG SATFDKGVSL 720
721 RFPRFLRIRE DKGVEDATSS DQIVELYENQ SHMQN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
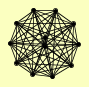
|
CAK1 CDC34 CDC53 CDC9 CRT10 GRR1 MEP3 MET30 MMS1 POR1 RTT101 SAF1 |
| View Details | Ho Y, et al. (2002) |
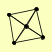
|
CDC9 DBP9 ECM10 POL30 YOR378W |
| View Details | Qiu et al. (2008) |
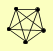
|
CDC9 DPB2 POL3 POL31 POL32 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
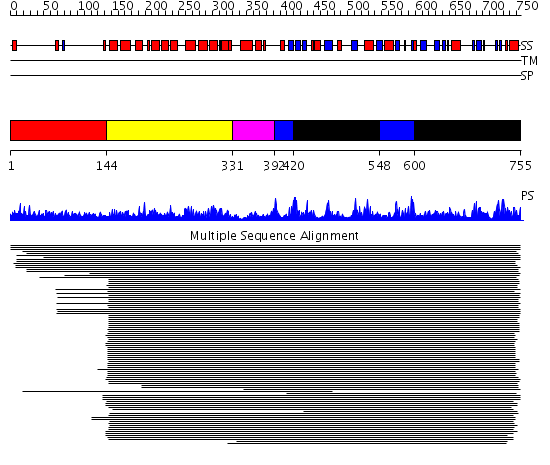
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 7.99 | No description for 1ldvA was found. | |
| 2 | View Details | [144..330] | 7.17 | Lipovitellin-phosvitin complex, superhelical domain; Lipovitellin-phosvitin complex; beta-sheet shell regions | |
| 3 | View Details | [331..391] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [392..419] [548..599] |
38.30103 | ATP-dependent DNA ligase; ATP-dependent DNA ligase, N-terminal domain | |
| 5 | View Details | [420..547] [600..755] |
38.30103 | ATP-dependent DNA ligase; ATP-dependent DNA ligase, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|