













| General Information: |
|
| Name(s) found: |
MMS1 /
YPR164W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1407 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
DNA repair
[IGI negative regulation of transposition, DNA-mediated [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLGLRTHGLD RYEHYIRRPS DFGKLELQDW LNHKSFRVSP NLLIDSSTTR EWNEPELFYQ 60
61 NTEDETWVRP CVGPKLEPSM MMLRYHDSNI GQMPQFCYPI SSPINFKPVL KYILQERSEL 120
121 SDGFPQKYNT LIGSLFDIDK NPETLDDSDI EALDDIEMSS DSGNVKEPKI ELQALEEIQQ 180
181 KHFSLIVSNN GIFQTGSTSI TYIQSGISGS IAIKPNNVAI LILLTQPSGH LLSILPLDDG 240
241 KETYLLQYWN LGQKGQWNII KHQNEKQFVL IHKELGICKF FEFHLPFTFQ LVNNLTLTDS 300
301 VIMNGSFFPT NYTDLDPYFI IFITAIRYER IVYFVIEWNN NEIKKKEVYQ LTVFDGEKTN 360
361 MTIPIGLNAC LVETPLKFSL VSANQIMSGE TEFHSFQLKA LKGIKSFFPA PLLLLKLQEL 420
421 HPHTFKKFQY CTIISSSTGN ICFCVTERST IVNGNLKFYE LTRFKGLKSI SPLPSNPINL 480
481 DSRSSSYVLV VISFSRTLEL TLSLEDLRCL DKKDVIKPLK NITFKHTIDS STEENSQILA 540
541 FTSSKFYNTH TGSNINDTRN SQVWLTSPNA ITQPCIDYKL RKTHQLIHLK QFQIFRHLRI 600
601 WKCKNLDIAL LQRLGINQSN TESSLIFATD AVSNNRIFLL DLTMTTTIDN DDPVQGLINI 660
661 EDLLCDTENE TILLNFTKNN LIQVTRDTIY IDPIGGDKEL RKISPGWEFE NVTYNDGILI 720
721 VWNAGLGCVS YIENIDAVDE SGALVSNLSS SKGMSKFFKQ LGTVTSVNFQ IKESTDDPTK 780
781 YDIWILLPDC VIRTPFSDWI SDSLDFSDVY ILSVQQALIN GPYFCSLDYE SYFEVHTLQN 840
841 NCFKKGSRCT SRVNFQGKDI KFRSFGVNQC LAFSAFEIFV INLTPIHDSR ELDFYKLKLP 900
901 HLGNNNSILE VCPDIENNQL FILYSDGLRI LELSYLTSNN GNFLLKSTRS KNKKFLYLDK 960
961 INRMLVLNQD LREWECIRLS DGKAVGLDSQ LLKDDSEEIL EIKELPIATE DNPLEKKTVL1020
1021 LISFTSSLKL VLLTAAKNKI SNQIIDSYKL DNSRLLNHLV ITPRGEIFFL DYKVMGTDNE1080
1081 MSFNKLKVTK HCIDQEERNN TTLRLTLETR FTFKSWSTVK TFTVVGDNII ATTNMGEKLY1140
1141 LIKDFSSSSD ESRRVYPLEM YPDSKVQKII PLNECCFVVA AYCGNRNDLD SRLIFYSLPT1200
1201 IKVGLNNETG SLPDEYGNGR VDDIFEVDFP EGFQFGTMAL YDVLHGERHV NRYSEGIRSE1260
1261 NDEAEVALRQ RRNLLLFWRN HSSTPKPSLR RAATIVYEDH VSSRYFEDIS SILGSTAMRT1320
1321 KRLSPYNAVA LDKPIQDISY DPAVQTLYVL MADQTIHKFG KDRLPCQDEY EPRWNSGYLV1380
1381 SRRSIVKSDL ICEVGLWNLS DNCKNTV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CAK1 CDC34 CDC53 CDC9 CRT10 GRR1 MEP3 MET30 MMS1 POR1 RTT101 SAF1 |
| View Details | Ho Y, et al. (2002) |

|
ADR1 BBC1 CDC39 CDC53 CRM1 DUR1,2 ECM29 ECM33 FAA4 GAL3 GCN1 HRT1 HYP2 MDN1 MKT1 MMS1 MYO2 PFK1 PMA2 RPA190 RPN1 TPS1 UBI4 VPS13 YAR009C YGP1 YLR035C-A |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
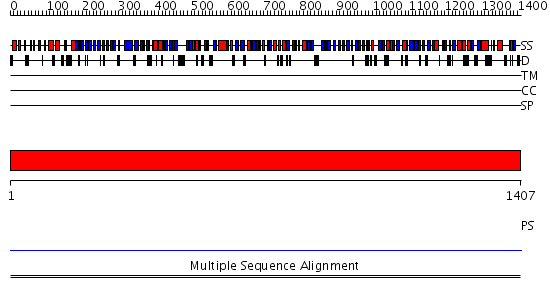
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1407] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [140..252] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [253..480] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [481..551] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [552..705] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [706..857] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [858..1061] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1062..1210] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1211..1407] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)