













| General Information: |
|
| Name(s) found: |
ADR1 /
YDR216W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1323 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
regulation of carbohydrate metabolic process
[IGI transcription [IGI peroxisome organization [IMP negative regulation of transcription from RNA polymerase II promoter by glucose [IMP |
| Molecular Function: |
transcription factor activity
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANVEKPNDC SGFPVVDLNS CFSNGFNNEK QEIEMETDDS PILLMSSSAS RENSNTFSVI 60
61 QRTPDGKIIT TNNNMNSKIN KQLDKLPENL RLNGRTPSGK LRSFVCEVCT RAFARQEHLK 120
121 RHYRSHTNEK PYPCGLCNRC FTRRDLLIRH AQKIHSGNLG ETISHTKKVS RTITKARKNS 180
181 ASSVKFQTPT YGTPDNGNFL NRTTANTRRK ASPEANVKRK YLKKLTRRAS FSAQSASSYA 240
241 LPDQSSLEQH PKDRVKFSTP ELVPLDLKNP ELDSSFDLNM NLDLNLNLDS NFNIALNRSD 300
301 SSGSTMNLDY KLPESANNYT YSSGSPTRAY VGANTNSKNA SFNDADLLSS SYWIKAYNDH 360
361 LFSVSESDET SPMNSELNDT KLIVPDFKST IHHLKDSRSS SWTVAIDNNS NNNKVSDNQP 420
421 DFVDFQELLD NDTLGNDLLE TTAVLKEFEL LHDDSVSATA TSNEIDLSHL NLSNSPISPH 480
481 KLIYKNKEGT NDDMLISFGL DHPSNREDDL DKLCNMTRDV QAIFSQYLKG EESKRSLEDF 540
541 LSTSNRKEKP DSGNYTFYGL DCLTLSKISR ALPASTVNNN QPSHSIESKL FNEPMRNMCI 600
601 KVLRYYEKFS HDSSESVMDS NPNLLSKELL MPAVSELNEY LDLFKNNFLP HFPIIHPSLL 660
661 DLDLDSLQRY TNEDGYDDAE NAQLFDRLSQ GTDKEYDYEH YQILSISKIV CLPLFMATFG 720
721 SLHKFGYKSQ TIELYEMSRR ILHSFLETKR RCRSTTVNDS YQNIWLMQSL ILSFMFALVA 780
781 DYLEKIDSSL MKRQLSALCS TIRSNCLPTI SANSEKSINN NNEPLTFGSP LQYIIFESKI 840
841 RCTLMAYDFC QFLKCFFHIK FDLSIKEKDV ETIYIPDNES KWASESIICN GHVVQKQNFY 900
901 DFRNFYYSFT YGHLHSIPEF LGSSMIYYEY DLRKGTKSHV FLDRIDTKRL ERSLDTSSYG 960
961 NDNMAATNKN IAILIDDTII LKNNLMSMRF IKQIDRSFTE KVRKGQIAKI YDSFLNSVRL1020
1021 NFLKNYSVEV LCEFLVALNF SIRNISSLYV EEESDCSQRM NSPELPRIHL NNQALSVFNL1080
1081 QGYYYCFILI IKFLLDFEAT PNFKLLRIFI ELRSLANSIL LPTLSRLYPQ EFSGFPDVVF1140
1141 TQQFINKDNG MLVPGLSANE HHNGASAAVK TKLAKKINVE GLAMFINEIL VNSFNDTSFL1200
1201 NMEDPIRNEF SFDNGDRAVT DLPRSAHFLS DTGLEGINFS GLNDSHQTVS TLNLLRYGEN1260
1261 HSSKHKNGGK GQGFAEKYQL SLKYVTIAKL FFTNVKENYI HCHMLDKMAS DFHTLENHLK1320
1321 GNS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |

|
ACC1 ACH1 ADH2 ADR1 ARC15 ARC18 ARC19 ARC35 ARC40 ARP2 ARP3 ATP3 AYR1 BBC1 BGL2 BMH1 BNI1 BNR1 BOI2 CBP6 CCT3 CDC39 CDC53 COP1 CPA2 CRM1 CSR2 CYK3 DEF1 DOP1 DUR1,2 ECM29 ECM33 ENP1 FAA1 FAA4 FOL2 GAL3 GAL7 GAR1 GCD11 GCN1 GFA1 GND1 GSP1 GSY2 GUS1 HEM15 HRP1 HRT1 HYP2 IDH1 IDH2 IMH1 ISA2 KAP104 KAP122 KCS1 KTR3 LSC1 LST8 LYS12 MCM4 MCM7 MDH1 MDN1 MET18 MKT1 MMS1 MRT4 MYO2 NAB2 NDE1 NGL2 NTH1 OAC1 PET10 PET9 PFK1 PHO86 PMA2 POR1 PRB1 PSD1 PYC1 PYC2 REG1 RET1 RET2 RPA135 RPA190 RPA49 RPB5 RPC19 RPC25 RPC34 RPC40 RPC82 RPN1 RPN12 RPN3 RPO26 RPO31 RPT4 RRP3 RVS167 SEC21 SEC26 SEC27 SEC28 SIP1 SLC1 SOK1 SRY1 STI1 STU1 SVL3 TBS1 TFG2 TFP1 TPS1 TRP3 TSA2 UBI4 VMA8 VPS13 YAR009C YBT1 YDL063C YER138C YFL042C YFR017C YGP1 YHM2 YHR112C YIL028W YJR029W YKU80 YLR035C-A YML020W YNL035C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
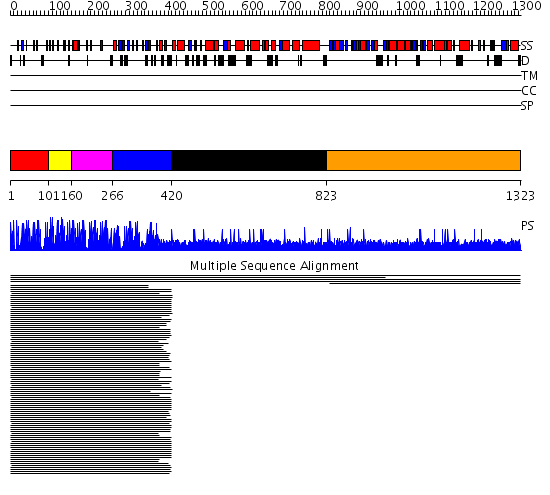
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | 112.440336 | ZIF268 | |
| 2 | View Details | [101..159] | 328.741366 | ADR1 | |
| 3 | View Details | [160..265] | 58.29073 | Ying-yang 1 (yy1, zinc finger domain) | |
| 4 | View Details | [266..419] | 7.692996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [420..822] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [823..1323] | 1.077 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)