













| General Information: |
|
| Name(s) found: |
KAP104 /
YBR017C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 918 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
mRNA-binding (hnRNP) protein import into nucleus
[IDA protein import into nucleus [IDA cell cycle [IMP |
| Molecular Function: |
nuclear localization sequence binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASTWKPAED YVLQLATLLQ NCMSPNPEIR NNAMEAMENF QLQPEFLNYL CYILIEGESD 60
61 DVLKQHYSLQ DLQNNRATAG MLLKNSMLGG NNLIKSNSHD LGYVKSNIIH GLYNSNNNLV 120
121 SNVTGIVITT LFSTYYRQHR DDPTGLQMLY QLLELTSNGN EPSIKALSKI MEDSAQFFQL 180
181 EWSGNTKPME ALLDSFFRFI SNPNFSPVIR SESVKCINTV IPLQTQSFIV RLDKFLEIIF 240
241 QLAQNDENDL VRAQICISFS FLLEFRPDKL VSHLDGIVQF MLHLITTVNE EKVAIEACEF 300
301 LHAFATSPNI PEHILQPYVK DIVPILLSKM VYNEESIVLL EASNDDDAFL EDKDEDIKPI 360
361 APRIVKKKEA GNGEDADDNE DDDDDDDDED GDVDTQWNLR KCSAATLDVM TNILPHQVMD 420
421 IAFPFLREHL GSDRWFIREA TILALGAMAE GGMKYFNDGL PALIPFLVEQ LNDKWAPVRK 480
481 MTCWTLSRFS PWILQDHTEF LIPVLEPIIN TLMDKKKDVQ EAAISSVAVF IENADSELVE 540
541 TLFYSQLLTS FDKCLKYYKK KNLIILYDAI GRFAEKCALD ETAMQIILPP LIEKWALLSD 600
601 SDKELWPLLE CLSCVASSLG ERFMPMAPEV YNRAFRILCH CVELEAKSHQ DPTIVVPEKD 660
661 FIITSLDLID GLVQGLGAHS QDLLFPQGTK DLTILKIMLE CLQDPVHEVR QSCFALLGDI 720
721 VYFFNSELVI GNLEDFLKLI GTEIMHNDDS DGTPAVINAI WALGLISERI DLNTYIIDMS 780
781 RIILDLFTTN TQIVDSSVME NLSVTIGKMG LTHPEVFSSG AFANDSNWNK WCLSVNALDD 840
841 VEEKSSAYMG FLKIINLTST EVTMSNDTIH KIVTGLSSNV EANVFAQEIY TFLMNHSAQI 900
901 SAINFTPDEI SFLQQFTS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ENP1 GEA2 HRP1 KAP104 KTR3 MIR1 NAB2 YHR020W YNL035C |
| View Details | Krogan NJ, et al. (2006) |

|
ALD4 KAP104 TAF6 TDA5 |
| View Details | Gavin AC, et al. (2006) |
|
CLU1 HRP1 KAP104 |
| View Details | Gavin AC, et al. (2002) |

|
BMH1 BMH2 CLU1 CRM1 ECM29 FKS1 GCN1 GEA2 GFA1 HRP1 KAP104 KAP123 LCB1 LCB2 LYS12 MIR1 NAB2 SAM1 SAM2 YHR020W |
| View Details | Ho Y, et al. (2002) |

|
ADH4 ADR1 ARC1 AYR1 BGL2 CBP6 DIM1 DPS1 ENP1 FAA1 GAR1 GSP1 GSP2 HEM15 HRP1 HTA2 KAP104 KTR3 MRT4 NAB2 NDE1 OAC1 PAA1 PET10 PET9 PGM2 PSD1 RPT4 RTN2 SAC1 TFG2 VMA8 YDL063C YHM2 YNL035C YRB1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
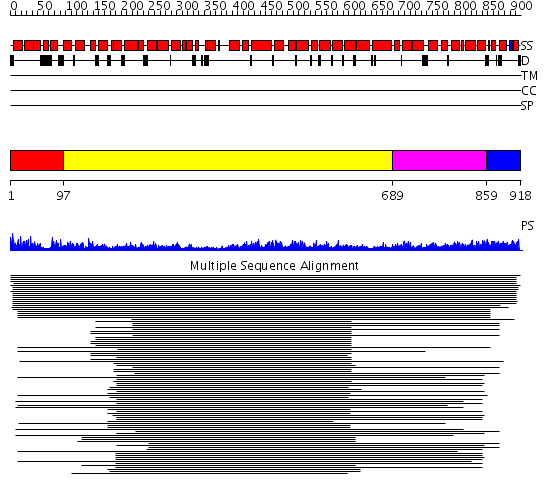
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [97..688] | 20.431997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [689..858] | 2.262988 | View MSA. Confident ab initio structure predictions are available. | |
| 4 | View Details | [859..918] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)