













| General Information: |
|
| Name(s) found: |
KAP123 /
YER110C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1113 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA nuclear pore [IDA |
| Biological Process: |
protein import into nucleus
[TAS |
| Molecular Function: |
protein transmembrane transporter activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQQFLSQLE QTLHAITSGV GLKEATKTLQ TQFYTQPTTL PALIHILQNG SDDSLKQLAG 60
61 VEARKLVSKH WNAIDESTRA SIKTSLLQTA FSEPKENVRH SNARVIASIG TEELDGNKWP 120
121 DLVPNLIQTA SGEDVQTRQT AIFILFSLLE DFTSSLSGHI DDFLALFSQT INDPSSLEIR 180
181 SLSAQALNHV SALIEEQETI NPVQAQKFAA SIPSVVNVLD AVIKADDTMN AKLIFNCLND 240
241 FLLLDSQLTG NFIVDLIKLS LQIAVNSEID EDVRVFALQF IISSLSYRKS KVSQSKLGPE 300
301 ITVAALKVAC EEIDVDDELN NEDETGENEE NTPSSSAIRL LAFASSELPP SQVASVIVEH 360
361 IPAMLQSANV FERRAILLAI SVAVTGSPDY ILSQFDKIIP ATINGLKDTE PIVKLAALKC 420
421 IHQLTTDLQD EVAKFHEEYL PLIIDIIDSA KNIVIYNYAT VALDGLLEFI AYDAIAKYLD 480
481 PLMNKLFYML ESNESSKLRC AVVSAIGSAA FAAGSAFIPY FKTSVHYLEK FIQNCSQIEG 540
541 MSEDDIELRA NTFENISTMA RAVRSDAFAE FAEPLVNSAY EAIKTDSARL RESGYAFIAN 600
601 LAKVYGENFA PFLKTILPEI FKTLELDEYQ FNFDGDAEDL AAFADSANEE ELQNKFTVNT 660
661 GISYEKEVAS AALSELALGT KEHFLPYVEQ SLKVLNEQVD ESYGLRETAL NTIWNVVKSV 720
721 LLASKVEPES YPKGIPASSY VNADVLAVIQ AARETSMGNL SDEFETSMVI TVMEDFANMI 780
781 KQFGAIIIMD NGDSSMLEAL CMQVLSVLKG THTCQTIDIE EDVPRDEELD ASETEATLQD 840
841 VALEVLVSLS QALAGDFAKV FDNFRPVVFG LFQSKSKNKR SSAVGAASEL ALGMKEQNPF 900
901 VHEMLEALVI RLTSDKSLEV RGNAAYGVGL LCEYASMDIS AVYEPVLKAL YELLSAADQK 960
961 ALAAEDDEAT REIIDRAYAN ASGCVARMAL KNSALVPLEQ TVPALLAHLP LNTGFEEYNP1020
1021 IFELIMKLYQ ENSPVITNET PRIIEIFSAV FTKENDRIKL EKESTLGREE NMERLKQFQT1080
1081 EEMKHKVIEL LKYLNTTYNG IVAQNPVLAA VIA |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
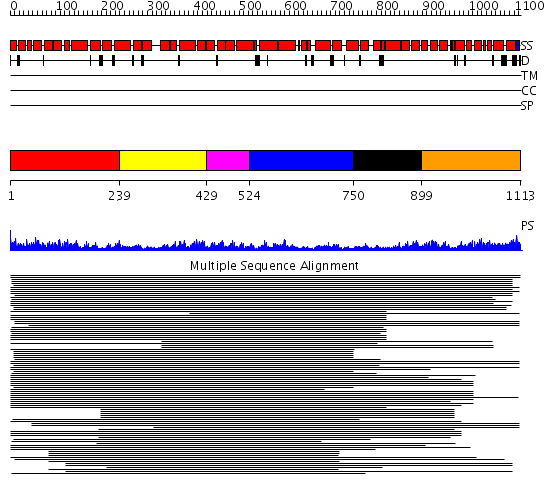
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..238] | 99.228787 | Importin beta | |
| 2 | View Details | [239..428] | 99.228787 | Importin beta | |
| 3 | View Details | [429..523] | 99.228787 | Importin beta | |
| 4 | View Details | [524..749] | 99.228787 | Importin beta | |
| 5 | View Details | [750..898] | 14.16 | Adaptin beta subunit N-terminal fragment | |
| 6 | View Details | [899..1113] | 14.16 | Adaptin beta subunit N-terminal fragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)