













| General Information: |
|
| Name(s) found: |
MEC1 /
YBR136W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2368 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC mitochondrion [IDA |
| Biological Process: |
DNA damage checkpoint
[TAS DNA replication checkpoint [IMP reciprocal meiotic recombination [IMP DNA recombination [IMP telomere maintenance [IDA telomere maintenance via recombination [IGI nucleobase, nucleoside, nucleotide and nucleic acid metabolic process [IGI |
| Molecular Function: |
protein kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESHVKYLDE LILAIKDLNS GVDSKVQIKK VPTDPSSSQE YAKSLKILNT LIRNLKDQRR 60
61 NNIMKNDTIF SKTVSALALL LEYNPFLLVM KDSNGNFEIQ RLIDDFLNIS VLNYDNYHRI 120
121 WFMRRKLGSW CKACVEFYGK PAKFQLTAHF ENTMNLYEQA LTEVLLGKTE LLKFYDTLKG 180
181 LYILLYWFTS EYSTFGNSIA FLDSSLGFTK FDFNFQRLIR IVLYVFDSCE LAALEYAEIQ 240
241 LKYISLVVDY VCNRTISTAL DAPALVCCEQ LKFVLTTMHH FLDNKYGLLD NDPTMAKGIL 300
301 RLYSLCISND FSKCFVDHFP IDQWADFSQS EHFPFTQLTN KALSIVYFDL KRRSLPVEAL 360
361 KYDNKFNIWV YQSEPDSSLK NVTSPFDDRY KQLEKLRLLV LKKFNKTERG TLLKYRVNQL 420
421 SPGFFQRAGN DFKLILNEAS VSIQTCFKTN NITRLTSWTV ILGRLACLES EKFSGTLPNS 480
481 TKDMDNWYVC HLCDIEKTGN PFVRINPNRP EAAGKSEIFR ILHSNFLSHP NIDEFSESLL 540
541 SGILFSLHRI FSHFQPPKLT DGNGQINKSF KLVQKCFMNS NRYLRLLSTR IIPLFNISDS 600
601 HNSEDEHTAT LIKFLQSQKL PVVKENLVIA WTQLTLTTSN DVFDTLLLKL IDIFNSDDYS 660
661 LRIMMTLQIK NMAKILKKTP YQLLSPILPV LLRQLGKNLV ERKVGFQNLI ELLGYSSKTI 720
721 LDIFQRYIIP YAIIQYKSDV LSEIAKIMCD GDTSLINQMK VNLLKKNSRQ IFAVALVKHG 780
781 LFSLDILETL FLNRAPTFDK GYITAYLPDY KTLAEITKLY KNSVTKDASD SENANMILCS 840
841 LRFLITNFEK DKRHGSKYKN INNWTDDQEQ AFQKKLQDNI LGIFQVFSSD IHDVEGRTTY 900
901 YEKLRVINGI SFLIIYAPKK SIISALAQIS ICLQTGLGLK EVRYEAFRCW HLLVRHLNDE 960
961 ELSTVIDSLI AFILQKWSEF NGKLRNIVYS ILDTLIKEKS DLILKLKPYT TLALVGKPEL1020
1021 GILARDGQFA RMVNKIRSTT DLIPIFANNL KSSNKYVINQ NLDDIEVYLR RKQTERSIDF1080
1081 TPKKVGQTSD ITLVLGALLD TSHKFRNLDK DLCEKCAKCI SMIGVLDVTK HEFKRTTYSE1140
1141 NEVYDLNDSV QTIKFLIWVI NDILVPAFWQ SENPSKQLFV ALVIQESLKY CGLSSESWDM1200
1201 NHKELYPNEA KLWEKFNSVS KTTIYPLLSS LYLAQSWKEY VPLKYPSNNF KEGYKIWVKR1260
1261 FTLDLLKTGT TENHPLHVFS SLIREDDGSL SNFLLPYISL DIIIKAEKGT PYADILNGII1320
1321 IEFDSIFTCN LEGMNNLQVD SLRMCYESIF RVFEYCKKWA TEFKQNYSKL HGTFIIKDTK1380
1381 TTNMLLRIDE FLRTTPSDLL AQRSLETDSF ERSALYLEQC YRQNPHDKNQ NGQLLKNLQI1440
1441 TYEEIGDIDS LDGVLRTFAT GNLVSKIEEL QYSENWKLAQ DCFNVLGKFS DDPKTTTRML1500
1501 KSMYDHQLYS QIISNSSFHS SDGKISLSPD VKEWYSIGLE AANLEGNVQT LKNWVEQIES1560
1561 LRNIDDREVL LQYNIAKALI AISNEDPLRT QKYIHNSFRL IGTNFITSSK ETTLLKKQNL1620
1621 LMKLHSLYDL SFLSSAKDKF EYKSNTTILD YRMERIGADF VPNHYILSMR KSFDQLKMNE1680
1681 QADADLGKTF FTLAQLARNN ARLDIASESL MHCLERRLPQ AELEFAEILW KQGENDRALK1740
1741 IVQEIHEKYQ ENSSVNARDR AAVLLKFTEW LDLSNNSASE QIIKQYQDIF QIDSKWDKPY1800
1801 YSIGLYYSRL LERKKAEGYI TNGRFEYRAI SYFLLAFEKN TAKVRENLPK VITFWLDIAA1860
1861 ASISEAPGNR KEMLSKATED ICSHVEEALQ HCPTYIWYFV LTQLLSRLLH SHQSSAQIIM1920
1921 HILLSLAVEY PSHILWYITA LVNSNSSKRV LRGKHILEKY RQHSQNPHDL VSSALDLTKA1980
1981 LTRVCLQDVK SITSRSGKSL EKDFKFDMNV APSAMVVPVR KNLDIISPLE SNSMRGYQPF2040
2041 RPVVSIIRFG SSYKVFSSLK KPKQLNIIGS DGNIYGIMCK KEDVRQDNQY MQFATTMDFL2100
2101 LSKDIASRKR SLGINIYSVL SLREDCGILE MVPNVVTLRS ILSTKYESLK IKYSLKSLHD2160
2161 RWQHTAVDGK LEFYMEQVDK FPPILYQWFL ENFPDPINWF NARNTYARSY AVMAMVGHIL2220
2221 GLGDRHCENI LLDIQTGKVL HVDFDCLFEK GKRLPVPEIV PFRLTPNLLD ALGIIGTEGT2280
2281 FKKSSEVTLA LMRKNEVALM NVIETIMYDR NMDHSIQKAL KVLRNKIRGI DPQDGLVLSV2340
2341 AGQTETLIQE ATSEDNLSKM YIGWLPFW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
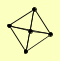
|
BZZ1 LAS17 MEC1 SLA1 VRP1 |
| View Details | Gavin AC, et al. (2006) |

|
MEC1 VAC14 |
| View Details | Gavin AC, et al. (2002) |
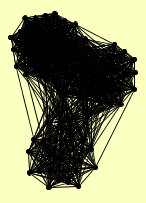
|
CLU1 CMR1 CRN1 DNA2 ECM29 HDA1 HDA2 HDA3 HSC82 KAP123 KAP95 LCD1 MEC1 MGM101 MIP6 MIS1 MLH1 MPH1 MRPL23 MRPS28 MSH2 MSH3 MSH6 NAB3 NAB6 NRD1 OLA1 PGK1 PMS1 PRO2 RAD16 RAD23 RAD34 RAD4 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 ROM2 RPA135 RPN10 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT6 RSC8 RVB2 SEN54 SGS1 SHS1 SRO7 SSL1 SUI3 SUP35 SUP45 TAO3 TFA1 TFB1 TFB4 TIF1, TIF2 TOP3 USO1 UTP20 VAC14 VAS1 VPS1 VPS5 YHR122W YJL017W |
| View Details | Ho Y, et al. (2002) |
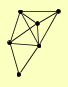
|
ACO1 CLU1 GUS1 MDH1 MEC1 STI1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
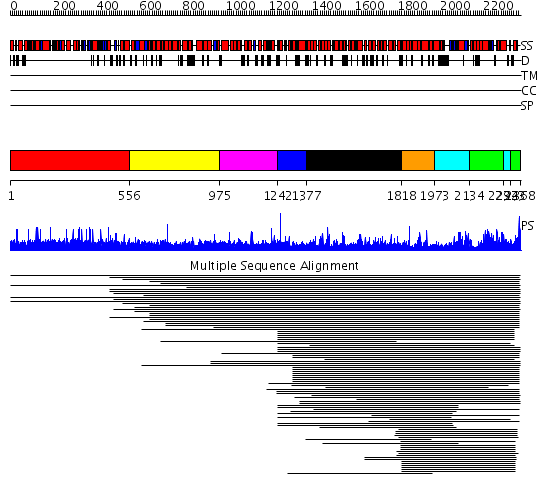
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..555] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [556..974] | 1.018981 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [975..1241] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1242..1376] | 1.040997 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [1377..1817] | 68.026872 | FAT domain No confident structure predictions are available. | |
| 6 | View Details | [1818..1972] | 166.9897 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 7 | View Details | [1973..2133] [2294..2322] |
166.9897 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 8 | View Details | [2134..2293] [2323..2368] |
166.9897 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 9 | View Details | [1644..1820] | 5.69897 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 10 | View Details | [1821..2368] | 105.0 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)