













| General Information: |
|
| Name(s) found: |
SUP45 /
YBR143C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 437 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IPI translation release factor complex [IDA |
| Biological Process: |
translational termination
[IMP cytokinesis [IGI |
| Molecular Function: |
translation release factor activity, codon specific
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNEVEKNIE IWKVKKLVQS LEKARGNGTS MISLVIPPKG QIPLYQKMLT DEYGTASNIK 60
61 SRVNRLSVLS AITSTQQKLK LYNTLPKNGL VLYCGDIITE DGKEKKVTFD IEPYKPINTS 120
121 LYLCDNKFHT EVLSELLQAD DKFGFIVMDG QGTLFGSVSG NTRTVLHKFT VDLPKKHGRG 180
181 GQSALRFARL REEKRHNYVR KVAEVAVQNF ITNDKVNVKG LILAGSADFK TDLAKSELFD 240
241 PRLACKVISI VDVSYGGENG FNQAIELSAE ALANVKYVQE KKLLEAYFDE ISQDTGKFCY 300
301 GIDDTLKALD LGAVEKLIVF ENLETIRYTF KDAEDNEVIK FAEPEAKDKS FAIDKATGQE 360
361 MDVVSEEPLI EWLAANYKNF GATLEFITDK SSEGAQFVTG FGGIGAMLRY KVNFEQLVDE 420
421 SEDEYYDEDE GSDYDFI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
SUP35 SUP45 |
| View Details | Krogan NJ, et al. (2006) |
|
PLB3 RPS10B SCM3 SUP35 SUP45 TRM12 VPS3 |
| View Details | Gavin AC, et al. (2006) |

|
SUP35 SUP45 |
| View Details | Gavin AC, et al. (2002) |
|
ASC1 ATG9 CLU1 CRN1 ECM29 FUN12 GLT1 HCR1 IST2 KAP123 KTR5 LSG1 MEC1 MIP6 MIS1 MRPL23 NAB3 NAB6 NIP1 NOG1 NOP7 NRD1 PRT1 RIC1 RLI1 ROM2 RPA135 RPG1 SEN54 SPT5 SUA7 SUI1 SUI2 SUI3 SUP35 SUP45 TAO3 TIF34 TIF35 TIF5 USO1 UTP20 UTP6 VAC14 VPS5 YGL036W YJL017W |
| View Details | Ho Y, et al. (2002) |
|
AAC3 CYS4 GND1 GSP1 MLC1 MYO4 NOP12 PUF6 RLI1 RPF2 SHE3 SPT2 SRP1 SSF1 STE23 SUL2 SUP45 TIF4631 UTP22 UTP7 |
| View Details | Qiu et al. (2008) |

|
SUP35 SUP45 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
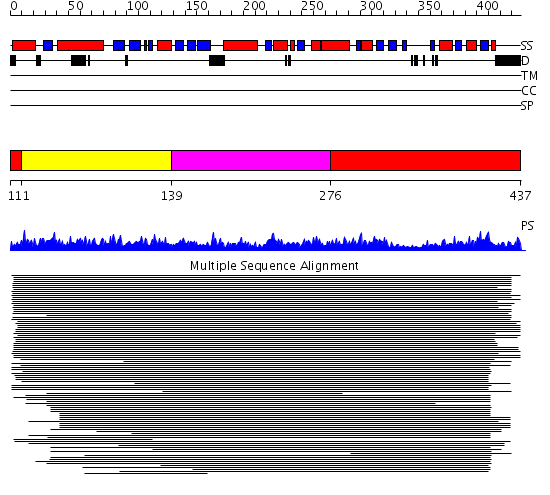
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..10] [276..437] |
2710.0 | Middle domain of eukaryotic peptide chain release factor subunit 1, ERF1; C-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1; N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 | |
| 2 | View Details | [11..138] | 2710.0 | Middle domain of eukaryotic peptide chain release factor subunit 1, ERF1; C-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1; N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 | |
| 3 | View Details | [139..275] | 2710.0 | Middle domain of eukaryotic peptide chain release factor subunit 1, ERF1; C-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1; N-terminal domain of eukaryotic peptide chain release factor subunit 1, ERF1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)