













| General Information: |
|
| Name(s) found: |
SUL2 /
YLR092W
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 893 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IMP |
| Biological Process: |
sulfate transport
[IMP |
| Molecular Function: |
sulfate transmembrane transporter activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSREGYPNFE EVEIPDFQET NNTVPDLDDL ELEYDQYKNN ENNDTFNDKD LESNSVAKHN 60
61 AVNSSKGVKG SKIDYFNPSD VSLYDNSVSQ FEETTVSLKE YYDHSIRSHL TVKGACSYLK 120
121 SVFPIINWLP HYNFSWFTAD LIAGITIGCV LVPQSMSYAQ VATLPAQYGL YSSFIGAYSY 180
181 SFFATSKDVC IGPVAVMSLQ TAKVIADVTA KYPDGDSAIT GPVIATTLAL LCGIISAAVG 240
241 FLRLGFLVEL ISLNAVAGFM TGSAFNILWG QVPALMGYNS LVNTRAATYK VVIETLKHLP 300
301 DTKLDAVFGL IPLFLLYVWK WWCGTYGPRL NDRYNSKNPR LHKIIKWTYF YAQASRNGII 360
361 IIVFTCIGWA ITRGKSKSER PISILGSVPS GLKEVGVFHV PPGLMSKLGP NLPASIIVLL 420
421 LEHIAISKSF GRINDYKVVP DQELIAIGVS NLLGTFFNAY PATGSFSRSA LKAKCNVRTP 480
481 LSGLFSGSCV LLALYCLTGA FFYIPKATLS AVIIHAVSDL LASYQTTWNF WKMNPLDFIC 540
541 FIVTVLITVF ASIEDGIYFA MCWSCAMLIL KVAFPAGKFL GRVEVAEVTD AYVRPDSDVV 600
601 SYVSENNNGI STLEDGGEDD KESSTKYVTN SSKKIETNVQ TKGFDSPSSS ISQPRIKYHT 660
661 KWIPFDHKYT RELNPDVQIL PPPDGVLVYR LSESYTYLNC SRHYNIITEE VKKVTRRGQL 720
721 IRHRKKSDRP WNDPGPWEAP AFLKNLKFWK KRENDPESME NAPSTSVDVE RDDRPLLKIL 780
781 CLDFSQVAQT DATALQSLVD LRKAINQYAD RQVEFHFVGI ISPWVKRGLI SRGFGTLNEE 840
841 YSDESIVAGH TSYHVARVPQ GEENPEKYSV YTASGTNLPF FHIDIPDFAK WDI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
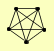
|
MLC1 MYO4 SHE3 SUL2 SUP45 |
| View Details | Qiu et al. (2008) |
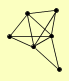
|
ARR3 PHO84 PHO89 SSU1 SUL1 SUL2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
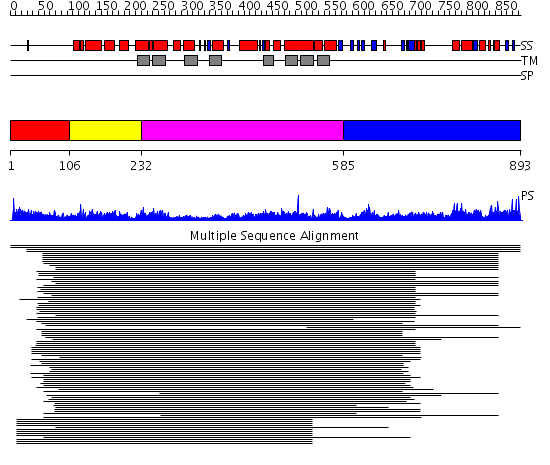
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 5.39794 | Erythrocite membrane Band 3 | |
| 2 | View Details | [106..231] | 2.379992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [232..584] | 119.79588 | Sulfate transporter family No confident structure predictions are available. | |
| 4 | View Details | [585..893] | 1.270961 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [710..893] | 1.31 | Solution Structure of putative anti sigma factor antagonist from Thermotoga maritima (TM1442) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|