













| General Information: |
|
| Name(s) found: |
SUL1 /
YBR294W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IMP |
| Biological Process: |
sulfate transport
[IGI |
| Molecular Function: |
sulfate transmembrane transporter activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRKSSTEYV HNQEDADIEV FESEYRTYRE SEAAENRDGL HNGDEENWKV NSSKQKFGVT 60
61 KNELSDVLYD SIPAYEESTV TLKEYYDHSI KNNLTAKSAG SYLVSLFPII KWFPHYNFTW 120
121 GYADLVAGIT VGCVLVPQSM SYAQIASLSP EYGLYSSFIG AFIYSLFATS KDVCIGPVAV 180
181 MSLQTAKVIA EVLKKYPEDQ TEVTAPIIAT TLCLLCGIVA TGLGILRLGF LVELISLNAV 240
241 AGFMTGSAFN IIWGQIPALM GYNSLVNTRE ATYKVVINTL KHLPNTKLDA VFGLIPLVIL 300
301 YVWKWWCGTF GITLADRYYR NQPKVANRLK SFYFYAQAMR NAVVIVVFTA ISWSITRNKS 360
361 SKDRPISILG TVPSGLNEVG VMKIPDGLLS NMSSEIPASI IVLVLEHIAI SKSFGRINDY 420
421 KVVPDQELIA IGVTNLIGTF FHSYPATGSF SRSALKAKCN VRTPFSGVFT GGCVLLALYC 480
481 LTDAFFFIPK ATLSAVIIHA VSDLLTSYKT TWTFWKTNPL DCISFIVTVF ITVFSSIENG 540
541 IYFAMCWSCA MLLLKQAFPA GKFLGRVEVA EVLNPTVQED IDAVISSNEL PNELNKQVKS 600
601 TVEVLPAPEY KFSVKWVPFD HGYSRELNIN TTVRPPPPGV IVYRLGDSFT YVNCSRHYDI 660
661 IFDRIKEETR RGQLITLRKK SDRPWNDPGE WKMPDSLKSL FKFKRHSATT NSDLPISNGS 720
721 SNGETYEKPL LKVVCLDFSQ VAQVDSTAVQ SLVDLRKAVN RYADRQVEFH FAGIISPWIK 780
781 RSLLSVKFGT TNEEYSDDSI IAGHSSFHVA KVLKDDVDYT DEDSRISTSY SNYETLCAAT 840
841 GTNLPFFHID IPDFSKWDV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
SSU1 SUL1 SUL2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
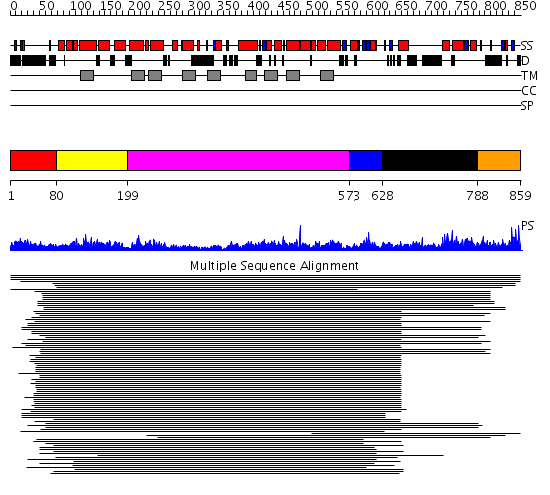
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..79] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [80..198] | 1.33299 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [199..572] | 101.0 | Sulfate transporter family No confident structure predictions are available. | |
| 4 | View Details | [573..627] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [628..787] | 3.086186 | STAS domain No confident structure predictions are available. | |
| 6 | View Details | [788..859] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|