













| General Information: |
|
| Name(s) found: |
PRT1 /
YOR361C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 763 amino acids |
Gene Ontology: |
|
| Cellular Component: |
multi-eIF complex
[IDA cytoplasm [IPI eukaryotic translation initiation factor 3 complex [IDA |
| Biological Process: |
translational initiation
[IDA |
| Molecular Function: |
translation initiation factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKNFLPRTLK NIYELYFNNI SVHSIVSRNT QLKRSKIIQM TTETFEDIKL EDIPVDDIDF 60
61 SDLEEQYKVT EEFNFDQYIV VNGAPVIPSA KVPVLKKALT SLFSKAGKVV NMEFPIDEAT 120
121 GKTKGFLFVE CGSMNDAKKI IKSFHGKRLD LKHRLFLYTM KDVERYNSDD FDTEFREPDM 180
181 PTFVPSSSLK SWLMDDKVRD QFVLQDDVKT SVFWNSMFNE EDSLVESREN WSTNYVRFSP 240
241 KGTYLFSYHQ QGVTAWGGPN FDRLRRFYHP DVRNSSVSPN EKYLVTFSTE PIIVEEDNEF 300
301 SPFTKKNEGH QLCIWDIASG LLMATFPVIK SPYLKWPLVR WSYNDKYCAR MVGDSLIVHD 360
361 ATKNFMPLEA KALKPSGIRD FSFAPEGVKL QPFRNGDEPS VLLAYWTPET NNSACTATIA 420
421 EVPRGRVLKT VNLVQVSNVT LHWQNQAEFL CFNVERHTKS GKTQFSNLQI CRLTERDIPV 480
481 EKVELKDSVF EFGWEPHGNR FVTISVHEVA DMNYAIPANT IRFYAPETKE KTDVIKRWSL 540
541 VKEIPKTFAN TVSWSPAGRF VVVGALVGPN MRRSDLQFYD MDYPGEKNIN DNNDVSASLK 600
601 DVAHPTYSAA TNITWDPSGR YVTAWSSSLK HKVEHGYKIF NIAGNLVKED IIAGFKNFAW 660
661 RPRPASILSN AERKKVRKNL REWSAQFEEQ DAMEADTAMR DLILHQRELL KQWTEYREKI 720
721 GQEMEKSMNF KIFDVQPEDA SDDFTTIEEI VEEVLEETKE KVE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
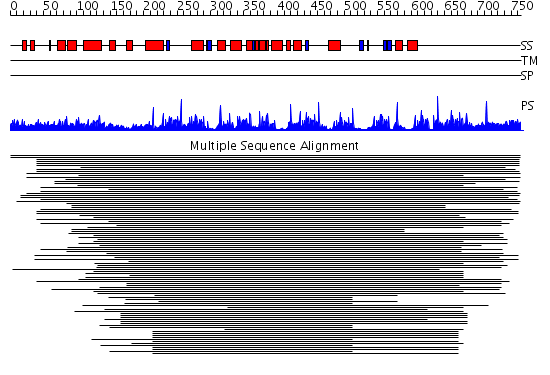
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | 16.69897 | Crystal Structure of UP1 Complexed With d(TAGG(6MI)TTAGGG): A Human Telomeric Repeat Containing 6-methyl-8-(2-deoxy-beta-ribofuranosyl)isoxanthopteridine (6MI) | |
| 2 | View Details | [161..679] | 37.522879 | Actin interacting protein 1 | |
| 3 | View Details | [680..763] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)