













| General Information: |
|
| Name(s) found: |
TSC11 /
YER093C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1430 amino acids |
Gene Ontology: |
|
| Cellular Component: |
TORC2 complex
[IPI |
| Biological Process: |
regulation of cell growth
[IPI cellular cell wall organization [IGI establishment or maintenance of actin cytoskeleton polarity [IMP sphingolipid biosynthetic process [IGI |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIPHSAKQS SPLSSRRRSV TNTTPLLTPR HSRDNSSTQI SSAKNITSSS PSTITNESSK 60
61 RNKQNLVLST SFISTKRLEN SAPSPTSPLM ARRTRSTMTK ALLNLKAEIN NQYQELARLR 120
121 KKKDDIEHLR DSTISDIYSG SYSTNHLQKH SMRIRANTQL REIDNSIKRV EKHIFDLKQQ 180
181 FDKKRQRSLT TSSSIKADVG SIRNDDGQNN DSEELGDHDS LTDQVTLDDE YLTTPTSGTE 240
241 RNSQQNLNRN STVNSRNNEN HSTLSIPDLD GSNKVNLTGD TEKDLGDLEN ENQIFTSTTT 300
301 EAATWLVSDY MQSFQEKNVN PDFIAQKANG LVTLLKEHSE IRKDLVLTSF MSSIQNLLLN 360
361 GNKLIAASAY RVCRYLINSS IFIDELLELR LDAFIIISLA KDNSFQIERE QALKMVRRFI 420
421 EYNNGVTQGI MQAIISCVEK PEDSLRHMAL ETLLELCFVA PEMVKECRGM RVIEGFLQDY 480
481 TSFSLASVIL DTILQLMATH KTRQHFLEDF NVSVLTTVFS DTNTKSNVNV EKMQNASTLI 540
541 SITLNSYNGF MLFSNNNFKP LKQLVSFFQI PICAQYLIDI FLDVLKIKPL PYKPRGRHSH 600
601 SFKPIPSQYY KECMSVNQRL ALIVLILENS EFVPHLLELL NEEDRDDHLV AKGRYLLTEY 660
661 FNLRMNLVDK KYTSVSKPIY KENFTYVNET FQFKKIAYKM NRNRNTIGMS GIDYAQNIKS 720
721 FSKNIKENTL LREVDDFRFR RMVYDSKVLQ TKDFTRWNWN IINELLEGPL LNKKQLEELV 780
781 KSTKFIRRLL VFYRPLRLRF SNVNKGAKLS QKYVQVGCQF FKTLTATPEG MKILMDDTKI 840
841 IPQLASLMFR AMEGNISGNI FNKNKLREKI IFGYFKFIGI LTQSKNGVHI LTRWNFFTVI 900
901 YKMFQFESKL GLEFLLLTIP ELDLKYSSHC RVIIGKALVV ANEKVRIEAT KHIGDKLKEL 960
961 LSTKESDLKL KANKVKLQQF KMEMLTRQLY DLSPSVVAVA DQALYECIVA GNGSEELGTS1020
1021 FRMFLNQMVF IRSPILFELL SRPYGFQLLN EINFVKEERD SWLSKKNIEY VHIVEEFLKK1080
1081 NESINAKSLT FQQKSRLPLH FYESLTKTED GILLLSQTGD LVTFMNVIKK YVNGNNMATV1140
1141 ENAKEILDLK AALWCVGFIG STELGIGLLD NYSLVEDIIE VAYNASVTSV RFTAFYVLGL1200
1201 ISMTREGCEI LDEMGWNCCV SVQDEPIGIA LPNRLDRFLS YNEHKWSAFG EYSDEMIVFN1260
1261 KSDGDLIEKC LPIEFDLDKL LKEKDTAENP LNEKIITNKY DNDITSQTIT VSGENSSLFA1320
1321 NEGLSSPYVT QYRNDDDSIE SKVLHIVSQL GNHILSNHAV KEITEINNKY GPRLFENEKM1380
1381 FFKVFNMMSK YRFKPHVRKF LCGLFINNRA LENVIRHDNK RDKRPANFTR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |

|
BDF2 CDC55 CKB1 DYN1 ECM5 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MDN1 MIS1 MSB1 NIP1 NMD5 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 TEL1 TSC11 USO1 UTP20 VTH2 YFR006W YPR077C |
| View Details | Ho Y, et al. (2002) |
|
AVO1 BEM3 CCT2 CYS4 DIG1 DIG2 GUS1 KSS1 NPA3 PIM1 RPA135 RPN6 RVB1 STE11 STE12 TEC1 TSC11 |
| View Details | Qiu et al. (2008) |
|
AVO1 AVO2 BIT61 LST8 SLM1 SLM2 TOR2 TSC11 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
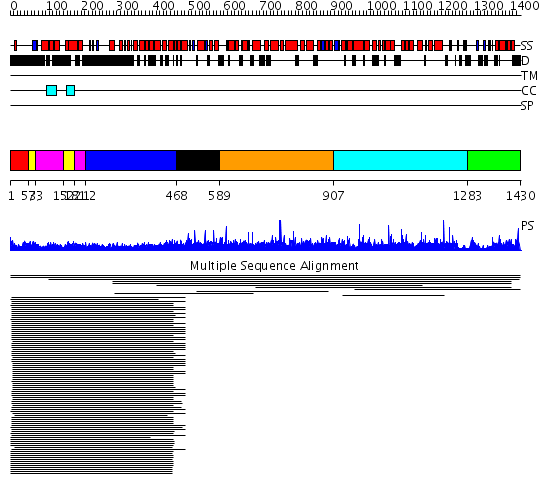
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..52] | 44.09691 | Heavy meromyosin subfragment | |
| 2 | View Details | [53..72] [152..180] |
5.154902 | Colicin Ia; Colicin Ia, N-terminal domain | |
| 3 | View Details | [73..151] [181..211] |
5.154902 | Colicin Ia; Colicin Ia, N-terminal domain | |
| 4 | View Details | [212..467] | 5.154902 | Colicin Ia; Colicin Ia, N-terminal domain | |
| 5 | View Details | [468..588] | 7.18 | Son of sevenless protein homolog 1 (sos-1) | |
| 6 | View Details | [589..906] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [907..1282] | 1.007917 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1283..1430] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1225..1299] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1300..1430] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)