













| General Information: |
|
| Name(s) found: |
RPN12 /
YFR052W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 274 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proteasome regulatory particle, lid subcomplex
[IPI |
| Biological Process: |
ubiquitin-dependent protein catabolic process
[IMP |
| Molecular Function: |
endopeptidase activity
[NAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSLAELTKS LSIAFENGDY AACEKLLPPI KIELIKNNLL IPDLSIQNDI YLNDLMITKR 60
61 ILEVGALASI QTFNFDSFEN YFNQLKPYYF SNNHKLSESD KKSKLISLYL LNLLSQNNTT 120
121 KFHSELQYLD KHIKNLEDDS LLSYPIKLDR WLMEGSYQKA WDLLQSGSQN ISEFDSFTDI 180
181 LKSAIRDEIA KNTELSYDFL PLSNIKALLF FNNEKETEKF ALERNWPIVN SKVYFNNQSK 240
241 EKADYEDEMM HEEDQKTNII EKAMDYAISI ENIV |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
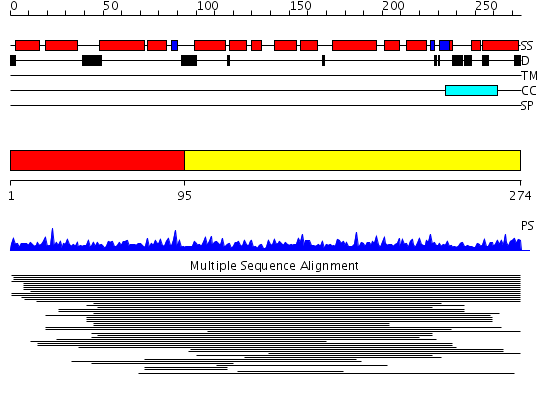
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [95..274] | 4.035977 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)