













| General Information: |
|
| Name(s) found: |
GFA1 /
YKL104C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 717 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
cell wall chitin biosynthetic process
[IGI |
| Molecular Function: |
glutamine-fructose-6-phosphate transaminase (isomerizing) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCGIFGYCNY LVERSRGEII DTLVDGLQRL EYRGYDSTGI AIDGDEADST FIYKQIGKVS 60
61 ALKEEITKQN PNRDVTFVSH CGIAHTRWAT HGRPEQVNCH PQRSDPEDQF VVVHNGIITN 120
121 FRELKTLLIN KGYKFESDTD TECIAKLYLH LYNTNLQNGH DLDFHELTKL VLLELEGSYG 180
181 LLCKSCHYPN EVIATRKGSP LLIGVKSEKK LKVDFVDVEF PEENAGQPEI PLKSNNKSFG 240
241 LGPKKAREFE AGSQNANLLP IAANEFNLRH SQSRAFLSED GSPTPVEFFV SSDAASVVKH 300
301 TKKVLFLEDD DLAHIYDGEL HIHRSRREVG ASMTRSIQTL EMELAQIMKG PYDHFMQKEI 360
361 YEQPESTFNT MRGRIDYENN KVILGGLKAW LPVVRRARRL IMIACGTSYH SCLATRAIFE 420
421 ELSDIPVSVE LASDFLDRKC PVFRDDVCVF VSQSGETADT MLALNYCLER GALTVGIVNS 480
481 VGSSISRVTH CGVHINAGPE IGVASTKAYT SQYIALVMFA LSLSDDRVSK IDRRIEIIQG 540
541 LKLIPGQIKQ VLKLEPRIKK LCATELKDQK SLLLLGRGYQ FAAALEGALK IKEISYMHSE 600
601 GVLAGELKHG VLALVDENLP IIAFGTRDSL FPKVVSSIEQ VTARKGHPII ICNENDEVWA 660
661 QKSKSIDLQT LEVPQTVDCL QGLINIIPLQ LMSYWLAVNK GIDVDFPRNL AKSVTVE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Dam1 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | sample 3928 - Asf1-S-TEV-ZZ (in hir3delta mutant) | Green EM, et al (2005) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
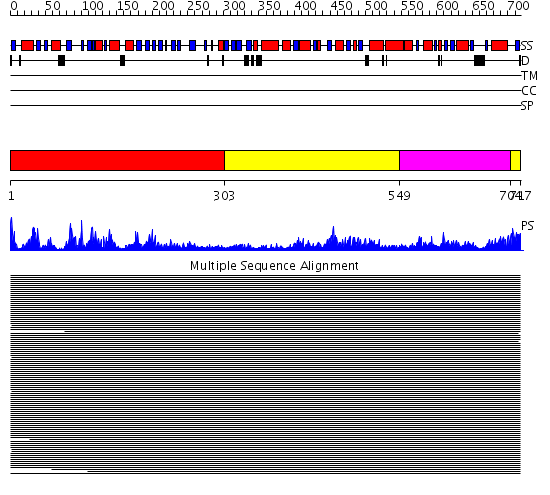
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..302] | 966.9897 | "Isomerase domain" of glucosamine 6-phosphate synthase (GLMS); Glucosamine 6-phosphate synthase, N-terminal domain | |
| 2 | View Details | [303..548] [704..717] |
966.9897 | "Isomerase domain" of glucosamine 6-phosphate synthase (GLMS); Glucosamine 6-phosphate synthase, N-terminal domain | |
| 3 | View Details | [549..703] | 966.9897 | "Isomerase domain" of glucosamine 6-phosphate synthase (GLMS); Glucosamine 6-phosphate synthase, N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)