













| General Information: |
|
| Name(s) found: |
YNL313C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 904 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
karyogamy involved in conjugation with cellular fusion
[IPI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METLLHAKLL LSAEVESLKS GSFDQTYVKK AEHIISGESY QLVQQFVDKF KGKISISGEI 60
61 STSSVIAALN DFLNVEVFKM GQENEMLFLA IALLQTFIQN NYTGPAARLK AISGLFGKTG 120
121 IEIGAVNTAL SRSLAIMGQP AYEFMDDPLY LVLSLLLLER ITGQKSLFDV TPDQEIPLPI 180
181 ISAESTPGLL AVAYWWWARA LLTQLSLIPE PSGFQASVAS AIYQSADLAY AITKELPESI 240
241 HEDFKRELCA MYYLENVKCS LAINTEHLCL PSLTRAKKIT NFEFVMTGAR ATRTKYQQKA 300
301 HAGLIILAKS FTFQNFALRT TSATPETFAL ESDLLLEKPH FESIADEPLD EQIYSKRQKV 360
361 DLNEGYEEDK LLPLALRQEN IPKLLLDLNP NDQPTLSDYD NIQLLLRLYT IKNTTPAKDP 420
421 LVEEELTALL SRILYQNGDK NWSIFARSLW ERSIIETTKA KTIERGLLQM QSLVEELDLK 480
481 IKSKLVPSSS EINVASRLSY IHQLPFIPRW QLDATLAEKY MSLGILKSAV EIYERLGMAC 540
541 ETALCYAAVG DEKKAEEILL QRINENDSDA RAYSILGDIK QDPSLWEKSW EIGKYVNAKN 600
601 SLAKYYFNPP PKSGAQPNYS ATLKHLNDSL RQYPLSFETW YFYGCVGLQC GKMQIAAEAF 660
661 TRCVSLDPYH ALSWSNLSAA YTKMDKLKEA YSCLKRAISC DAQKNWKIWE NYMLVAVKLN 720
721 EWEDVLTACK QLVSIRRDKS GEGSIDLPII EKLVELLVTS EYPEEPQQLS YFQKSCTEFI 780
781 CNTLPQVITT SARCWRLVAR VELWRKRPWA ALECHEKAYR AISHNPDLEV EEKVWNDTVD 840
841 ACEDLVAAYE SLGEMEGKYG PGSLVCKDWK YKCRSTIKAL MSKGKGRWDD SPGWDRLVEA 900
901 RSQI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Hazbun TR, et al. (2003) |
|
GRS1 KAR2 TUB3 YNL313C |
| View Details | Krogan NJ, et al. (2006) |

|
HSP26 YNL313C |
| View Details | Gavin AC, et al. (2002) |
|
ACE2 ALT2 ATP2 BCK1 CDC11 DRE2 ECM30 ENP2 GFA1 IML1 KAP123 LYS12 MNL1 MTR4 MYO2 RGC1 RPT5 SEC3 TFP1 UBP15 YBL044W YJR098C YNL313C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Unpublished Data |
| View Data |
|
Huh WK, et al. (2003) |
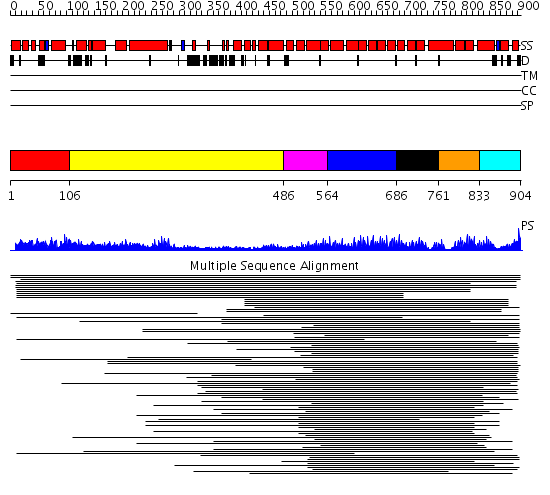
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [106..485] | 1.634996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [486..563] | 5.0 | Clathrin heavy chain proximal leg segment | |
| 4 | View Details | [564..685] | 38.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 5 | View Details | [686..760] | 38.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 6 | View Details | [761..832] | 38.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 7 | View Details | [833..904] | 38.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)