













| General Information: |
|
| Name(s) found: |
RGC1 /
YPR115W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1083 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDYFTFPKQ ENGGISKQPA TPGSTRSSSR NLELPKNYRS FGGSSDELAS MYSADSQYLM 60
61 DMIPDSLTLK NEPASGNTQM NGPDGKENKD IKLDEYILPK TDPRSPYYIN MPIPKKLPKS 120
121 EGKARAKQKV NRADPSDLDV ENIYETSGEF VREYPTDILI DRFHKWKKIL KSLIAYFREA 180
181 AYSQEQIARI NYQMKNAVKF AFLTDLEDET NKLVDPSISK LPTKKPQPVP LAAQKLDSKY 240
241 DTDVEQPQSI QSVPSEEVAS ASSGFMKFGS GSIQDIQVIL KKYHLSLGSQ QYKISKEILA 300
301 YIIPKLTDLR KDLTTKMKEI KELNGDFKTN IGEHIKITSR LLNKYIASVK LLDEASTSGD 360
361 KQGEKLKPKH DPYLLKLQLD LQLKRQLLEE NYLREAFLNL QSAALQLEKI VYSKIQSALQ 420
421 RYSALIDSEA RLMIKNLCHE LQQGILSRPP AVEWDNFVSH HPTCLMNLKS TDPPPQPRRL 480
481 SDIVYPNMKS PLAKCIRVGY LLKKTESSKS FTKGYFVLTT NYLHEFKSSD FFLDSKSPRS 540
541 KNKPVVEQSD ISRVNKDGTN AGSHPSSKGT QDPKLTKRRK GLSSSNLYPI SSLSLNDCSL 600
601 KDSTDSTFVL QGYASYHSPE DTCTKESSTT SDLACPTKTL ASNKGKHQRT PSALSMVSVP 660
661 KFLKSSSVPK EQKKAKEEAN INKKSICEKR VEWTFKIFSA SLEPTPEESK NFKKWVQDIK 720
721 ALTSFNSTQE RSNFIEEKIL KSRNHNNGKS SQRSKNSTYI TPVDSFVNLS EKVTPSSSVT 780
781 TLNTRKRANR PRYIDIPKSA NMNAGAMNSV YRSKVNTPAI DENGNLAIVG ETKNSAPQNG 840
841 MSYTIRTPCK SPYSPYTGEG MLYNRSADNL MASSSRKASA PGEVPQIAVS NHGDEAIIPA 900
901 SAYSDSSHKS SRASSVASIH NQRVDFYPSP LMNLPGVSPS CLALDGNANG YFGIPLNCNS 960
961 EARRGSDLSP FEMESPLFEE NRTQNCSGSR KSSACHIPHQ CGPRKEGNDS RLIYGNEKGA1020
1021 SQSRLTLKEP LTSKGVEAPY SSLKKTYSAE NVPLTSTVSN DKSLHSRKEG STNTVPATSA1080
1081 SSK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
AEP3 CDC20 FAB1 FOL2 GZF3 LSP1 NOP13 PIL1 PKH1 PRP16 PSK2 RGC1 RPL9B YLR179C YMR086W YTA6 |
| View Details | Gavin AC, et al. (2002) |
|
ACE2 ALT2 BCK1 CDC11 DRE2 ECM30 ENP2 IML1 MNL1 MTR4 MYO2 RGC1 SEC3 UBP15 YBL044W YJR098C YNL313C |
| View Details | Ho Y, et al. (2002) |

|
ARP7 DIG1 DIG2 FET4 GFA1 GUS1 HAS1 HXT6 KSS1 MKT1 MSE1 NAP1 PHO84 PIM1 PMA1 PYC1 RGC1 RPA135 RPN10 SEN1 STE12 STE7 TEC1 UBI4 YDR239C YHR033W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
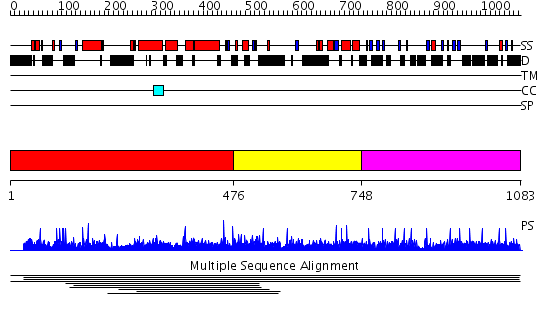
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..475] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [476..747] | 6.958607 | PH domain No confident structure predictions are available. | |
| 3 | View Details | [748..1083] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [469..805] | 0.982 | Insulin receptor substrate 1, IRS-1 | |
| 5 | View Details | [806..966] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [967..1083] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 | No functions predicted. | ||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 |
|
||||||||||||
| 5 | No functions predicted. | ||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)