













| General Information: |
|
| Name(s) found: |
NAP1 /
YKR048C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 417 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI cytoplasm [IDA nuclear nucleosome [IDA |
| Biological Process: |
M phase of mitotic cell cycle
[IGI budding cell bud growth [IGI nucleosome assembly [IDA |
| Molecular Function: |
histone binding
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDPIRTKPK SSMQIDNAPT PHNTPASVLN PSYLKNGNPV RAQAQEQDDK IGTINEEDIL 60
61 ANQPLLLQSI QDRLGSLVGQ DSGYVGGLPK NVKEKLLSLK TLQSELFEVE KEFQVEMFEL 120
121 ENKFLQKYKP IWEQRSRIIS GQEQPKPEQI AKGQEIVESL NETELLVDEE EKAQNDSEEE 180
181 QVKGIPSFWL TALENLPIVC DTITDRDAEV LEYLQDIGLE YLTDGRPGFK LLFRFDSSAN 240
241 PFFTNDILCK TYFYQKELGY SGDFIYDHAE GCEISWKDNA HNVTVDLEMR KQRNKTTKQV 300
301 RTIEKITPIE SFFNFFDPPK IQNEDQDEEL EEDLEERLAL DYSIGEQLKD KLIPRAVDWF 360
361 TGAALEFEFE EDEEEADEDE DEEEDDDHGL EDDDGESAEE QDDFAGRPEQ APECKQS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | control 1 | McCusker D, et al (2007) | |
| View Run | control 2 | McCusker D, et al (2007) | |
| View Run | experimental1 - mih1-1 | McCusker D, et al (2007) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Uetz P, et al. (2000) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
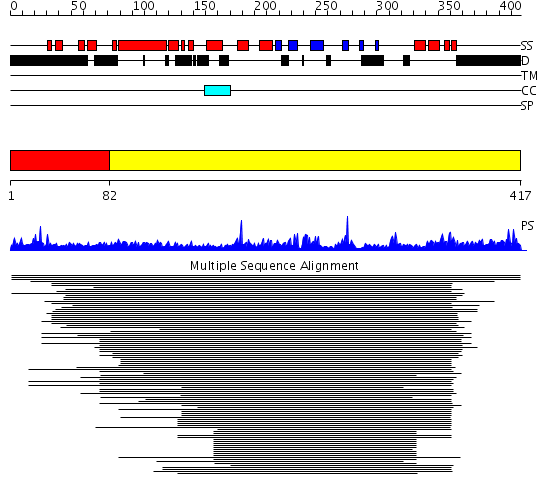
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [82..417] | 151.30103 | Nucleosome assembly protein (NAP) No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)