













| General Information: |
|
| Name(s) found: |
COS111 /
YBR203W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 924 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
signal transduction
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSASRLQNV NIVSNNYSRY GTSVYDKLYH SNGSGSNNAG KNSTTVGKLS SISQKSRSKQ 60
61 RHGSNCSRSM SQSPLSTFKS PLSNQNQSSA PDDLASIGQR RSDDVTSLDN ETIITMNSRK 120
121 SRIKKKYKSL ISTSSKKFMN KLYDHGASSD SFSIFSLKTS HSGKHENSRF EKLRKRKYHA 180
181 WGKFADINDL PVEIIAKILS EFELGRDQKT LVRCLYVSKK FYKATKIVLY RLPYFTSTYR 240
241 VAQFVTSLRL HPDNGAYVKV LDLSHLKPGI IGQDSKDSQG LDDDGHSRRH RRRRRRSTNT 300
301 SLNLPPATPT STISNEDDAN SGLIKDDASN GSEVEDLALA GWRDWRYRNE PLYSSPLLNS 360
361 FKLKKVVSRS SSITSTSSGN STGVHSTRRQ RSNSSVASIT TSIMSSIYNT SHVSLSSTTS 420
421 NTSNGNISSG SNLSRVSTAG SLKKASAKST RSSPQKAKPI SDITSSSWFR MRLSSRNRKA 480
481 RTANTINLKN SKDKSDDDFK VLKHDSGHPS NYRSSTLKFS IEQPFSTHHP YANKFLLKYA 540
541 PYKDLPLGYI LHMLNLCPNL VELNLSNLVI CTDFKLINQR SERRRMTSSL LPAVQESSVS 600
601 AGPEKDLEIV YMTDSGKGYE YYEGLSKKHS RSSSLGTNPS SWIGGQANWT DYPPPIDAQT 660
661 KTREEHRRNN TLNNKNVVLK KLNPFEIFEM ICNRNEEKGG YCSLTKVKMN DIVWCRQYMV 720
721 KYFVMRSWRN DLDYKSMENN SYERHLFSFR DSGLDRNFSW ACNAKLHEFV ALMVMDHLSN 780
781 LDDLGLEELF NIKSEKLYIK NYCCRDPDIL EISNLFDIRY GAGSEADATS DSNLEAESLQ 840
841 FRLTILKTEK PTSFWLTKVS KDYVSLVVKL CVDDDIDMDK MKVGKPTLRI DSITHNLISR 900
901 LKELRRVDLR RNVGENSYYA GSII |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
COS111 NAP1 SKP1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
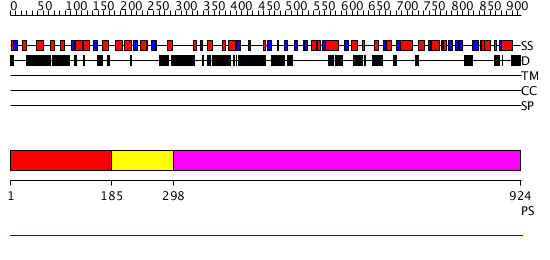
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [185..297] | 2.638272 | F-box domain Confident ab initio structure predictions are available. | |
| 3 | View Details | [298..924] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)