













| General Information: |
|
| Name(s) found: |
NIP1 /
YMR309C
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 812 amino acids |
Gene Ontology: |
|
| Cellular Component: |
multi-eIF complex
[IDA cytoplasm [IDA eukaryotic translation initiation factor 3 complex [IDA |
| Biological Process: |
ribosome biogenesis
[RCA translational initiation [IDA |
| Molecular Function: |
translation initiation factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRFFSSNYE YDVASSSSEE DLLSSSEEDL LSSSSSESEL DQESDDSFFN ESESESEADV 60
61 DSDDSDAKPY GPDWFKKSEF RKQGGGSNKF LKSSNYDSSD EESDEEDGKK VVKSAKEKLL 120
121 DEMQDVYNKI SQAENSDDWL TISNEFDLIS RLLVRAQQQN WGTPNIFIKV VAQVEDAVNN 180
181 TQQADLKNKA VARAYNTTKQ RVKKVSRENE DSMAKFRNDP ESFDKEPTAD LDISANGFTI 240
241 SSSQGNDQAV QEDFFTRLQT IIDSRGKKTV NQQSLISTLE ELLTVAEKPY EFIMAYLTLI 300
301 PSRFDASANL SYQPIDQWKS SFNDISKLLS ILDQTIDTYQ VNEFADPIDF IEDEPKEDSD 360
361 GVKRILGSIF SFVERLDDEF MKSLLNIDPH SSDYLIRLRD EQSIYNLILR TQLYFEATLK 420
421 DEHDLERALT RPFVKRLDHI YYKSENLIKI METAAWNIIP AQFKSKFTSK DQLDSADYVD 480
481 NLIDGLSTIL SKQNNIAVQK RAILYNIYYT ALNKDFQTAK DMLLTSQVQT NINQFDSSLQ 540
541 ILFNRVVVQL GLSAFKLCLI EECHQILNDL LSSSHLREIL GQQSLHRISL NSSNNASADE 600
601 RARQCLPYHQ HINLDLIDVV FLTCSLLIEI PRMTAFYSGI KVKRIPYSPK SIRRSLEHYD 660
661 KLSFQGPPET LRDYVLFAAK SMQKGNWRDS VKYLREIKSW ALLPNMETVL NSLTERVQVE 720
721 SLKTYFFSFK RFYSSFSVAK LAELFDLPEN KVVEVLQSVI AELEIPAKLN DEKTIFVVEK 780
781 GDEITKLEEA MVKLNKEYKI AKERLNPPSN RR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
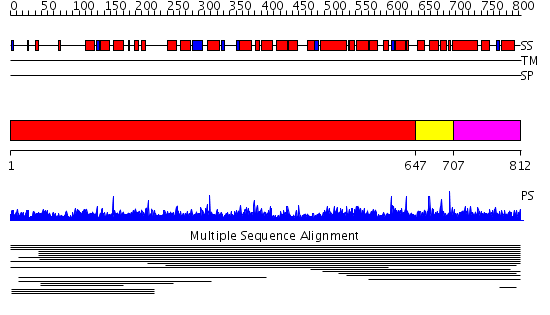
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..646] | 1.014001 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [647..706] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [707..812] | 15.420216 | PCI domain No confident structure predictions are available. | |
| 4 | View Details | [358..442] | 1.040983 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [443..535] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [536..662] | 1.048959 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [663..812] | 1.9 | Solution structure of the PCI domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)