













| General Information: |
|
| Name(s) found: |
TEL1 /
YBL088C
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2787 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC mitochondrion [IDA |
| Biological Process: |
DNA double-strand break processing
[IGI telomere maintenance [IMP telomere maintenance via telomerase [IMP phosphorylation [IMP response to DNA damage stimulus [IMP |
| Molecular Function: |
protein kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDHGIVETL NFLSSTKIKE RNNALDELTT ILKEDPERIP TKALSTTAEA LVELLASEHT 60
61 KYCDLLRNLT VSTTNKLSLS ENRLSTISYV LRLFVEKSCE RFKVKTLKLL LAVVPELMVK 120
121 DGSKSLLDAV SVHLSFALDA LIKSDPFKLK FMIHQWISLV DKICEYFQSQ MKLSMVDKTL 180
181 TNFISILLNL LALDTVGIFQ VTRTITWTVI DFLRLSKKEN GNTRLIMSLI NQLILKCHCF 240
241 SVIDTLMLIK EAWSYNLTIG CTSNELVQDQ LSLFDVMSSE LMNHKLPYMI GQENYVEELR 300
301 SESLVSLYRE YILLRLSNYK PQLFTVNHVE FSYIRGSRDK NSWFALPDFR LRDRGGRSVW 360
361 LKILGITKSL LTYFALNRKN ENYSLLFKRR KCDSDIPSIL RISDDMDTFL IHLLEENSSH 420
421 EFEVLGLQLC SFYGTLQDFT KSFAEQLKEL LFSKFEKIQC FNWVCFSFIP LLSQKECELS 480
481 NGDMARLFKV CLPLVKSNES CQLSCLLLAN SIKFSKQLLS DEKTINQIYD LYELSDILGP 540
541 ILVTNESFML WGYLQYVGKD FQSMNGISSA DRIFEWLKSK WNQLRGTDAK QDQFCNFISW 600
601 LGNKYDPENP FNDKKGEGAN PVSLCWDESH KIWQHFQEQR EFLLGVKPEE KSECFNTPFF 660
661 NLPKVSLDLT RYNEILYRLL ENIESDAFSS PLQKFTWVAK LIQIVDNLCG DSTFSEFIAA 720
721 YKRTTLITIP QLSFDSQNSY QSFFEEVLSI RTINVDHLVL DKINMKEIVN DFIRMQKNKS 780
781 QTGTSAINYF EASSEDTTQN NSPYTIGGRF QKPLHSTIDK AVRAYLWSSR NKSISERLVA 840
841 ILEFSDCVST DVFISYLGTV CQWLKQAIGE KSSYNKILEE FTEVLGEKLL CNHYSSSNQA 900
901 MLLLTSYIEA IRPQWLSYPE QPLNSDCNDI LDWIISRFED NSFTGVAPTV NLSMLLLSLL 960
961 QNHDLSHGSI RGGKQRVFAT FIKCLQKLDS SNIINIMNSI SSYMAQVSYK NQSIIFYEIK1020
1021 SLFGPPQQSI EKSAFYSLAM SMLSLVSYPS LVFSLEDMMT YSGFNHTRAF IQQALNKITV1080
1081 AFRYQNLTEL FEYCKFDLIM YWFNRTKVPT SKLEKEWDIS LFGFADIHEF LGRYFVEISA1140
1141 IYFSQGFNQK WILDMLHAIT GNGDAYLVDN SYYLCIPLAF ISGGVNELIF DILPQISGKT1200
1201 TVKYHKKYRL LMLKWIIRFT DLGSLTELRS TVEKLFPTSY LSPYLFENSS VSMRYQYPLH1260
1261 IPLALGATLV QTQFAHEKNN THEFKLLFLS VITDLEKTST YIGKLRCARE LKYLFVLYEN1320
1321 VLVKSSTLNF IIIRLSKFLI DTQIHDEVIT IFSSLLNLAD KNTFEIEPSL PNLFCKIFIY1380
1381 LRENKQLSPS FQQAIKLLEH RDLIKIKTWK YFLDAIFGNI VQDDIYENTE LLDASDCGVD1440
1441 DVVLVSLLFS YARRPVASKI GCSLSKAAAI NILKHHVPKE YLSKNFKLWF AALSRRILQQ1500
1501 EVQRERSTNF NNEVHLKNFE MVFRHPEQPH MIYQRISTFN KEAELYDSTE VFFISECILT1560
1561 YLVGYSIGNS ESEFCFRDNI MNENKDKVAP LDKDVLNAIY PLANNFGMES FICDTYLSVN1620
1621 EPYNCWLSKF ARSLIHQISF NIPPIVCLYP LCKGSTAFCE LVLTDLFFLS TTYDPKSCLN1680
1681 WSNRIFTQIA MLLHVKDSEI KLKMLFNVIK MIRMGSRCKE RNCLRIYSSL DLQEICQISL1740
1741 KIKEFKFGYL LFEEMNMPNI REMNINTLQK IYECINDGDF LAGLPVPHSI EGVLNSINRI1800
1801 DSDTWKRFLF NNADFDANYT TSLEEEKESL IKATEDSGFY GLTSLLESRL SGSSDVYKWN1860
1861 LELGDWKLLT PKVVDSKAKG LYYAIKNLPQ DVGFAEKSLE KSLLTIFDSR QHFISQTEWM1920
1921 DTLNAIIEFI KIAAIPQDVT SFPQTLMSIM KADKERLNTI DFYDHKTTLK SRHTLMNVLS1980
1981 RNSLDENVKC SKYLRLGSII QLANYVQLAI ANGAPQDALR NATLMSKTVK NIAKLYDDPS2040
2041 VVSQIEKLAS FTSANALWES REYKAPVMIM RDLLAQNEKN ISESILYDDF KLLINVPMDQ2100
2101 IKARLVKWSS ESRLEPAAAI YEKIIVNWDI NVEDHESCSD VFYTLGSFLD EQAQKLRSNG2160
2161 EIEDREHRSY TGKSTLKALE LIYKNTKLPE NERKDAKRHY NRVLLQYNRD SEVLKALLLQ2220
2221 KEKFLWHALH FYLNTLVFSN RYDNDIIDKF CGLWFENDDN SKINQLLYKE IGTIPSWKFL2280
2281 PWVNQIASKI SMEENEFQKP LQLTMKRLLY KLPYDSLYSV MSILLYEKQS NKDTNISQKI2340
2341 QAVKKILLEL QGYDRGAFAK KYLLPVQEFC EMSVELANLK FVQNTKTLRL ANLKIGQYWL2400
2401 KQLNMEKLPL PTSNFTVKSS ADGRKARPYI VSVNETVGIT TTGLSLPKIV TFNISDGTTQ2460
2461 KALMKGSNDD LRQDAIMEQV FQQVNKVLQN DKVLRNLDLG IRTYKVVPLG PKAGIIEFVA2520
2521 NSTSLHQILS KLHTNDKITF DQARKGMKAV QTKSNEERLK AYLKITNEIK PQLRNFFFDS2580
2581 FPDPLDWFEA KKTYTKGVAA SSIVGYILGL GDRHLNNILL DCSTGEPIHI DLGIAFDQGK2640
2641 LLPIPELVPF RLTRDIVDGF GVTGVDGLFR RSCERVYAVL RKDYVKVMCV LNILKWDPLY2700
2701 SWVMSPVKKY EHLFEEEHEI TNFDNVSKFI SNNDRNENQE SYRALKGVEE KLMGNGLSVE2760
2761 SSVQDLIQQA TDPSNLSVIY MGWSPFY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Gavin AC, et al. (2002) |

|
BDF2 CDC55 CKB1 DYN1 ECM5 FUN30 GLY1 HAT1 HAT2 HHF1, HHF2 HIF1 HSM3 ICL1 IML1 KAP123 KRE33 LIP2 MAC1 MDN1 MIS1 MSB1 NIP1 NMD5 PFK1 PRO1 PRT1 PSD2 PSE1 PSH1 RPA135 RPC40 RPG1 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT5 RPT6 RSC3 SAM1 TEL1 TSC11 USO1 UTP20 VTH2 YFR006W YPR077C |
| View Details | Ho Y, et al. (2002) |

|
GDE1 TEL1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
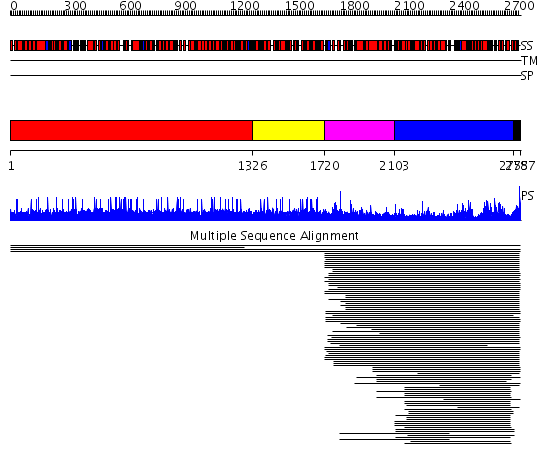
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..1325] | 1.002001 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [1326..1719] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [1720..2102] | 12.537602 | FAT domain No confident structure predictions are available. | |
| 4 | View Details | [2103..2754] | 167.980454 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain | |
| 5 | View Details | [2755..2787] | 15.920819 | FATC domain No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)