GO TERM SUMMARY
|
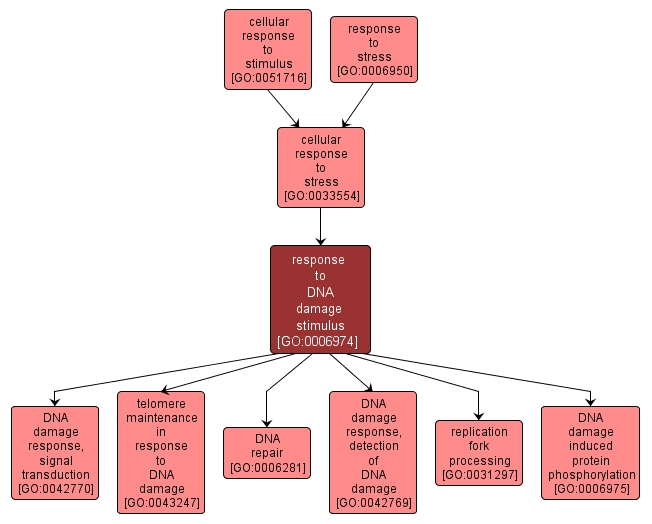
| Name: |
response to DNA damage stimulus |
| Acc: |
GO:0006974 |
| Aspect: |
Biological Process |
| Desc: |
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism. |
Synonyms:
- DNA damage response
- cellular DNA damage response
- GO:0034984
- cellular response to DNA damage stimulus
- response to genotoxic stress
|
|

|
INTERACTIVE GO GRAPH
|