













| General Information: |
|
| Name(s) found: |
RPN3 /
YER021W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 523 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proteasome regulatory particle, lid subcomplex
[IPI |
| Biological Process: |
ubiquitin-dependent protein catabolic process
[TAS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASTAVMMDV DSSGVNDLHH SEKKYAEEDQ VQELLKVLNE ISKTTLTLDP RYIWRSLKDL 60
61 SSLRNQELLN AETLCFTVNV LYPDSSSFKK NLLKFITSNH KSSVPGSAEL RNSYPASFYS 120
121 VNTEKKTIEV TAEINCFMHL LVQLFLWDSK ELEQLVEFNR KVVIPNLLCY YNLRSLNLIN 180
181 AKLWFYIYLS HETLARSSEE INSDNQNIIL RSTMMKFLKI ASLKHDNETK AMLINLILRD 240
241 FLNNGEVDSA SDFISKLEYP HTDVSSSLEA RYFFYLSKIN AIQLDYSTAN EYIIAAIRKA 300
301 PHNSKSLGFL QQSNKLHCCI QLLMGDIPEL SFFHQSNMQK SLLPYYHLTK AVKLGDLKKF 360
361 TSTITKYKQL LLKDDTYQLC VRLRSNVIKT GIRIISLTYK KISLRDICLK LNLDSEQTVE 420
421 YMVSRAIRDG VIEAKINHED GFIETTELLN IYDSEDPQQV FDERIKFANQ LHDEYLVSMR 480
481 YPEDKKTQQN EKSENGENDD DTLDGDLMDD MSDISDLDDL GFL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
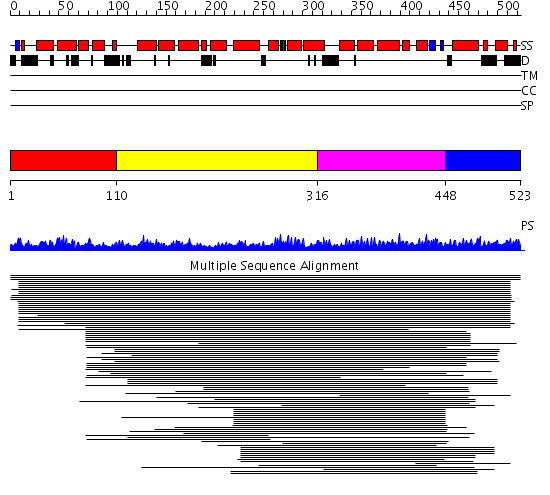
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 1.024992 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [110..315] | 10.38 | Transcription factor MalT domain III | |
| 3 | View Details | [316..447] | 10.38 | Transcription factor MalT domain III | |
| 4 | View Details | [448..523] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)