













| General Information: |
|
| Name(s) found: |
PTH2 /
YBL057C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 214 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IC mitochondrion [IDA mitochondrial outer membrane [IDA |
| Biological Process: |
translation
[IC negative regulation of proteasomal ubiquitin-dependent protein catabolic process [IDA |
| Molecular Function: |
aminoacyl-tRNA hydrolase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MITSFLMEKM TVSSNYTIAL WATFTAISFA VGYQLGTSNA SSTKKSSATL LRSKEMKEGK 60
61 LHNDTDEEES ESEDESDEDE DIESTSLNDI PGEVRMALVI RQDLGMTKGK IAAQCCHAAL 120
121 SCFRHIATNP ARASYNPIMT QRWLNAGQAK ITLKCPDKFT MDELYAKAIS LGVNAAVIHD 180
181 AGRTQIAAGS ATVLGLGPAP KAVLDQITGD LKLY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
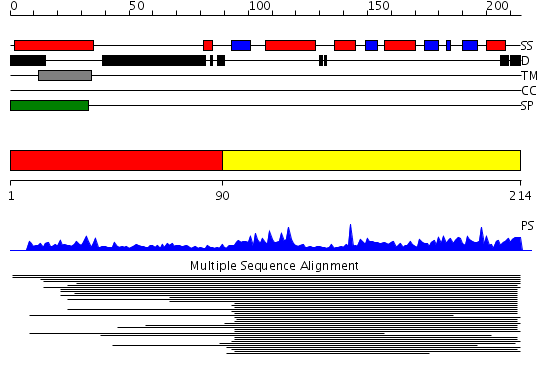
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [90..214] | 55.920819 | Peptidyl-tRNA hydrolase PTH2 No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|