













| General Information: |
|
| Name(s) found: |
ECM29 /
YHL030W
[SGD]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1868 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA proteasome complex [IDA |
| Biological Process: |
protein catabolic process
[IC |
| Molecular Function: |
protein binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSISSDEAKE KQLVEKAELR LAIADSPQKF ETNLQTFLPP LLLKLASPHA SVRTAVFSAL 60
61 KNLISRINTL PQVQLPVRAL IVQAKEPNLA AQQDSTNVRL YSLLLASKGI DRLSLQDRQQ 120
121 LLPLVVSNIS CLTGTVAARM FHILLKLILE WVAPQESSHE QEEFVQFLQL DNDGFSFLMR 180
181 QFTRFFLLVP SKQVQVSQQP LSRGYTCPGL SLTDVAFFTY DAGVTFNKEQ LNKFKKAIFQ 240
241 FVCRGMAATQ TIEQSPRMIE LMEFLCVVST DSTNLSDDAA QFMKRFPMPY ENEEFITFLQ 300
301 TLYIGNTANG RPPVKAILQE KILSILNRSH FATTKAECIS LICSIGLHSS EYKLRSLTLS 360
361 FIRHVAKLNY KNLNPASSSP SSTDFSTCIV SLIRNNLHAE GWPKLQLGPQ TPAFNTAILQ 420
421 RQLQYETLGD ILKRDFELVS DLSYIEFLFE SLKNDLPQFR SSIQESLLSL VGHLSILPQQ 480
481 SKLKLKNLLR KNLSIDEQQR EDNNDAVNSI MALKFVSIKF TNAAFPFHDP EARLFNIWGT 540
541 VRTNRFDIIE ESFKGLQPFW FRVNNASINT SATVKTSDLL GSHLSETEFP PFREFLQVLI 600
601 DQLDSEAASI TRKSLNNAVR FSKQCLISNA IYGKKTMVIQ DEDWSVRIDK ALELDDTVVS 660
661 RVNEMVQGMN DDIFIRYLTL LSNEFTATNS KGEQIAIFPY QDPIFGSVLL TLLNFVSNNV 720
721 LRRLEILVPD LYHLVIMKFQ SLSDNDLAVC ATIIGIISTA IADSTHVKRI TKIAQSQTMA 780
781 ETYVASYVVP RLYLKDQTNH IESDSILNLL NILTTHLSHP GTNKDMILKL VCQVTKFGLL 840
841 LQVSAQERKD FLKKVMDTIQ DKLINDVTAI QTWSYLSLYS TDLENSSLFQ EKLLETNVSK 900
901 QNDFLFSVGE SLSVVAGKWS SKYLIKQIDI PNFNVEIMQQ KFPATNVTTI LDEIFSGCDS 960
961 TKPSLRKASC IWLLSYIQYL GHLPEVSSKC NDIHLRFMRF LADRDEFIQD SAARGLSLVY1020
1021 EIGGSDLKES MVKGLLKSFT ESTAGSASTS ATGISGSVSE ETELFEPGVL NTGDGSISTY1080
1081 KDILNLASEV GDPALVYKFM SLAKSSALWS SRKGIAFGLG AIMSKSSLEE LLLKDQQTAK1140
1141 KLIPKLYRYR FDPFQAVSRS MTDIWNTLIP ESSLTISLYF NDILDELLCG MANKEWRVRE1200
1201 ASTSALLQLI QSQPQEKFSE KMLKIWTMAF RTMDDIKDSV REVGTKFTTV LAKILARSID1260
1261 VEKGVNPTKS KEILDNILPF LWGPHGLNSD AEEVRNFALT TLIDLVKHSP GAIKPFTPKL1320
1321 IYDFITLFSS IEPQVINYLA LNAANYNIDA NVIDTQRKNG VTNSPLFQTI EKLINNSDDC1380
1381 MMEEIINVVI KASRKSVGLP SKVASSLVII ILVKRYSIEM KPYSGKLLKV CLTMFEDRNE1440
1441 SVNIAFAISM GYLFKVSALD KCIKYSEKLI TKYFEPTSTE NNKKVVGTAI DSILNYAKSE1500
1501 FDNVASVFMP LIFIACNDED KDLETLYNKI WTEASSSGAG TVKLYLPEIL NVLCVNIKSN1560
1561 DFSIRKTCAK SVIQLCGGIN DSIPYPQIVK LFDISREALS GRSWDGKEHI VAALVSLTEK1620
1621 FSQTVADNND LQESINHVMY TEVSRKSMKY VKKILPLYAR YINVNPQEET ITFLIEKAKE1680
1681 MIRLLGSESD DSEGSIKQTS DESTIKRIKP NTEITQKSSK ENIENEEYVI NLLKVSVDIC1740
1741 NNSKSRYPMN LLEFIIDEIA YLFHNDRIIH TWRTQLAASE IGISIVGRFS TISSADFIQN1800
1801 VGRLWDQTFP INCNKETIEN VKLQMIKFGG LIIQKIPSLQ NNIEENLRLL NSIDSTSRIE1860
1861 LELKNIGL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
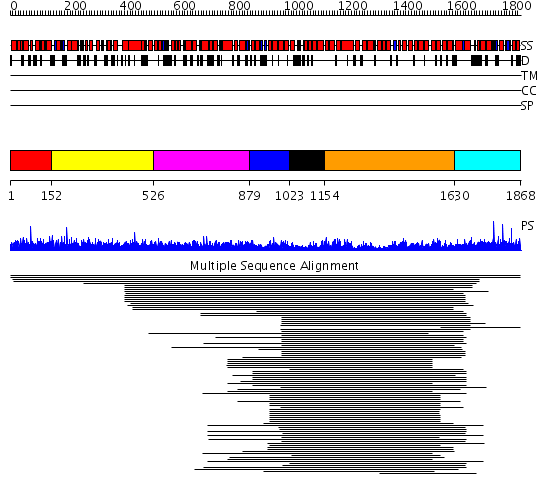
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 1.005972 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [152..525] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [526..878] | 3.083968 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [879..1022] | 46.0 | Importin beta | |
| 5 | View Details | [1023..1153] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1154..1629] | 19.377997 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1630..1868] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)