













| General Information: |
|
| Name(s) found: |
CDC39 /
YCR093W
[SGD]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 2108 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA CCR4-NOT core complex [IPI |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[TAS response to pheromone involved in conjugation with cellular fusion [IMP pseudohyphal growth [IMP nuclear-transcribed mRNA poly(A) tail shortening [IDA nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay [IDA regulation of transcription from RNA polymerase II promoter [IPI RNA elongation from RNA polymerase II promoter [IGI |
| Molecular Function: |
3'-5'-exoribonuclease activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSATYRDLN TASNLETSKE KQAAQIVIAQ ISLLFTTLNN DNFESVEREI RHILDRSSVD 60
61 IYIKVWERLL TLSSRDILQA GKFLLQENLL HRLLLEFAKD LPKKSTDLIE LLKERTFNNQ 120
121 EFQKQTGITL SLFIDLFDKS ANKDIIESLD RSSQINDFKT IKMNHTNYLR NFFLQTTPET 180
181 LESNLRDLLH SLEGESLNDL LALLLSEILS PGSQNLQNDP TRSWLTPPMV LDATNRGNVI 240
241 ARSISSLQAN QINWNRVFNL MSTKYFLSAP LMPTTASLSC LFAALHDGPV IDEFFSCDWK 300
301 VIFKLDLAIQ LHKWSVQNGC FDLLNAEGTR KVSETIPNTK QSLLYLLSIA SLNLELFLQR 360
361 EELSDGPMLA YFQECFFEDF NYAPEYLILA LVKEMKRFVL LIENRTVIDE ILITLLIQVH 420
421 NKSPSSFKDV ISTITDDSKI VDAAKIIINS DDAPIANFLK SLLDTGRLDT VINKLPFNEA 480
481 FKILPCARQI GWEGFDTFLK TKVSPSNVDV VLESLEVQTK MTDTNTPFRS LKTFDLFAFH 540
541 SLIEVLNKCP LDVLQLQRFE SLEFSLLIAF PRLINFGFGH DEAILANGDI AGINNDIEKE 600
601 MQNYLQKMYS GELAIKDVIE LLRRLRDSDL PRDQEVFTCI THAVIAESTF FQDYPLDALA 660
661 TTSVLFGSMI LFQLLRGFVL DVAFRIIMRF AKEPPESKMF KFAVQAIYAF RIRLAEYPQY 720
721 CKDLLRDVPA LKSQAQVYQS IVEAATLANA PKERSRPVQE MIPLKFFAVD EVSCQINQEG 780
781 APKDVVEKVL FVLNNVTLAN LNNKVDELKK SLTPNYFSWF STYLVTQRAK TEPNYHDLYS 840
841 KVIVAMGSGL LHQFMVNVTL RQLFVLLSTK DEQAIDKKHL KNLASWLGCI TLALNKPIKH 900
901 KNIAFREMLI EAYKENRLEI VVPFVTKILQ RASESKIFKP PNPWTVGILK LLIELNEKAN 960
961 WKLSLTFEVE VLLKSFNLTT KSLKPSNFIN TPEVIETLSG ALGSITLEQQ QTEQQRQIIL1020
1021 MQQHQQQMLI YQQRQQQQQQ RQQQQQHHIS ANTIADQQAA FGGEGSISHD NPFNNLLGST1080
1081 IFVTHPDLKR VFQMALAKSV REILLEVVEK SSGIAVVTTT KIILKDFATE VDESKLKTAA1140
1141 IIMVRHLAQS LARATSIEPL KEGIRSTMQS LAPNLMSLSS SPAEELDTAI NENIGIALVL1200
1201 IEKASMDKST QDLADQLMQA IAIRRYHKER RADQPFITQN TNPYSLSLPE PLGLKNTGVT1260
1261 PQQFRVYEEF GKNIPNLDVI PFAGLPAHAP PMTQNVGLTQ PQQQQAQMPT QILTSEQIRA1320
1321 QQQQQQLQKS RLNQPSQSAQ PPGVNVPNPQ GGIAAVQSDL EQNQRVLVHL MDILVSQIKE1380
1381 NATKNNLAEL GDQNQIKTII FQILTFIAKS AQKDQLALKV SQAVVNSLFA TSESPLCREV1440
1441 LSLLLEKLCS LSLVARKDVV WWLVYALDSR KFNVPVIRSL LEVNLIDATE LDNVLVTAMK1500
1501 NKMENSTEFA MKLIQNTVLS DDPILMRMDF IKTLEHLASS EDENVKKFIK EFEDTKIMPV1560
1561 RKGTKTTRTE KLYLVFTEWV KLLQRVENND VITTVFIKQL VEKGVISDTD NLLTFVKSSL1620
1621 ELSVSSFKES DPTDEVFIAI DALGSLIIKL LILQGFKDDT RRDYINAIFS VIVLVFAKDH1680
1681 SQEGTTFNER PYFRLFSNIL YEWATIRTHN FVRISDSSTR QELIEFDSVF YNTFSGYLHA1740
1741 LQPFAFPGFS FAWVTLLSHR MLLPIMLRLP NKIGWEKLML LIIDLFKFLD QYTSKHAVSD1800
1801 AVSVVYKGTL RVILGISNDM PSFLIENHYE LMNNLPPTYF QLKNVILSAI PKNMTVPNPY1860
1861 DVDLNMEDIP ACKELPEVFF DPVIDLHSLK KPVDNYLRIP SNSLLRTILS AIYKDTYDIK1920
1921 KGVGYDFLSV DSKLIRAIVL HVGIEAGIEY KRTSSNAVFN TKSSYYTLLF NLIQNGSIEM1980
1981 KYQIILSIVE QLRYPNIHTY WFSFVLMNMF KSDEWNDQKL EVQEIILRNF LKRIIVNKPH2040
2041 TWGVSVFFTQ LINNNDINLL DLPFVQSVPE IKLILQQLVK YSKKYTTSEQ DDQSATINRR2100
2101 QTPLQSNA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
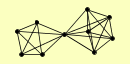
|
CAF130 CAF40 CCR4 CDC39 POP2 RPN1 RPN11 RPN2 RPN8 SPG5 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
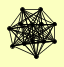
|
CAF16 CAF4 CCR4 CDC36 CDC39 DBF2 DHH1 IBA57 MOB1 MOT2 NOT3 NOT5 POP2 |
| View Details | Krogan NJ, et al. (2006) |
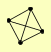
|
CAF40 CCR4 CDC39 POP2 |
| View Details | Gavin AC, et al. (2006) |
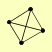
|
CAF40 CCR4 CDC39 POP2 |
| View Details | Gavin AC, et al. (2002) |

|
AHA1 ATP11 CAF130 CAF40 CCR4 CCT6 CDC36 CDC39 COP1 FAS2 GCN1 MOT2 NOT3 NOT5 PDC1 POL1 POL12 POP2 PRI1 PRI2 RVB2 SAM1 SEC27 TFC7 TFP1 YRA1 |
| View Details | Ho Y, et al. (2002) |
|
ADR1 AHP1 BBC1 CCR4 CDC36 CDC39 CDC53 CRM1 CYS4 DCS1 DCS2 DUR1,2 ECM29 ECM33 FAA4 GAL3 GCN1 GDB1 HRT1 HYP2 MDN1 MKT1 MMS1 MYO2 PFK1 PIL1 PMA2 POP2 RAD50 RNQ1 RPA190 RPN1 RVB1 STI1 TPS1 TSA2 UBI4 UBP15 UBR1 VPS1 VPS13 YAK1 YAR009C YGP1 YLR035C-A YLR241W YPL247C |
| View Details | Qiu et al. (2008) |
|
CAF16 CAF4 CAF40 CCR4 CDC36 CDC39 DBF2 DHH1 IBA57 MOB1 MOT2 NOT3 NOT5 POP2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Widlund PO, et al. (2005) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
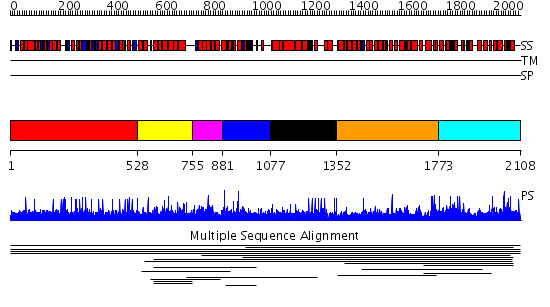
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..527] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [528..754] | 1.008947 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [755..880] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [881..1076] | 1.008956 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1077..1351] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1352..1772] | 1.010898 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1773..2108] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1079..1286] | 1.046996 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1287..1382] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1383..1457] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1458..1522] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1523..1709] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1710..1862] | N/A | No confident structure predictions are available. | |
| 14 | View Details | [1863..2108] | 1.058925 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.32 |
Source: Reynolds et al. (2008)