













| General Information: |
|
| Name(s) found: |
AHP1 /
YLR109W
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 176 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
response to oxidative stress
[IGI response to metal ion [IMP |
| Molecular Function: |
thioredoxin peroxidase activity
[IDA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
AHP1 HYP2 |
| View Details | Krogan NJ, et al. (2006) |
|
AHP1 MRT4 SSB2 |
| View Details | Ho Y, et al. (2002) |

|
AAC3 AAT2 ACH1 ACO1 ADH2 AFR1 AHA1 AHP1 ARC1 ARF1 ATP3 CAR2 CDC16 CDC33 CDC39 CLU1 COF1 COR1 CPR1 CYR1 CYS4 DCS1 DCS2 DNL4 DUN1 EGD1 ERG13 ERG20 ERG6 FOL2 FPR1 FRS2 GCD11 GDB1 GND1 GND2 GPG1 GRX1 GUS1 HEF3 HHF1, HHF2 HHT1, HHT2 HTB1 HXT7 HYP2 ILV5 LHS1 LIF1 LRO1 LSP1 MEC3 MET6 MGE1 MGT1 MNP1 MPC54 MRE11 NMD3 NTF2 OLA1 OYE2 OYE3 PFK1 PIL1 POR1 PRM2 PST2 PYC2 RAD50 RFA2 RGI1 RIP1 RNR2 RPP0 RSN1 RTC3 SAH1 SCP160 SEC27 SEC53 SEN15 SES1 SNU13 SOD1 SOF1 TEF4 THS1 TIF1, TIF2 TIF6 TMA19 TSA2 UBA1 UBC7 UBI4 UBP15 UBR1 VMA4 VMA5 VPS1 WTM1 XRS2 YAK1 YBL108W YCK1 YHR033W YHR112C YLR241W YNK1 YPL247C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #08 Alpha Factor Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
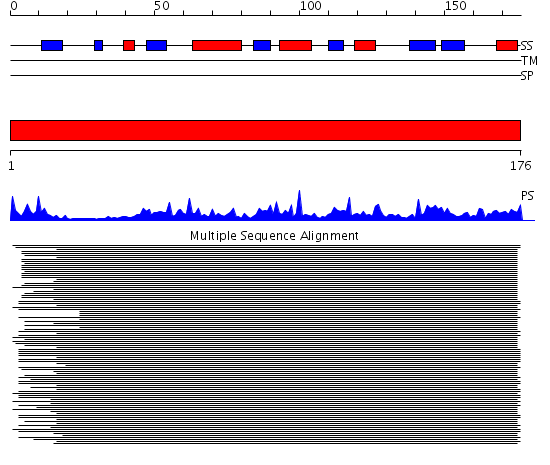
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | 121.770868 | Peroxiredoxin 5 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)